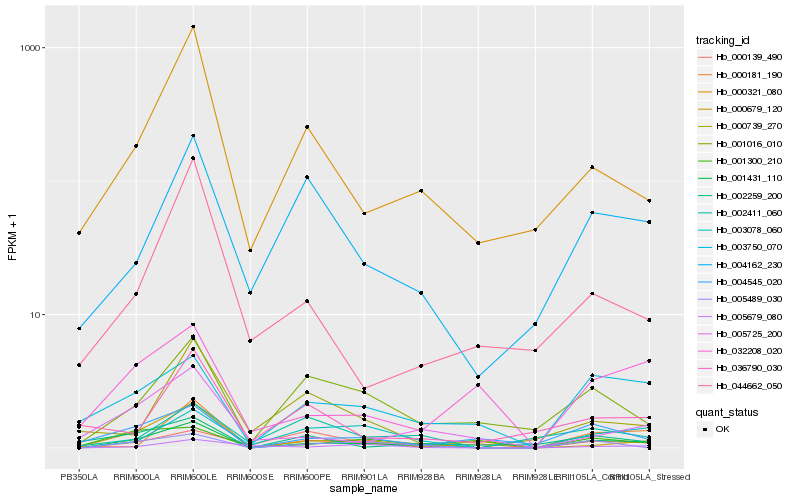
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001431_110 |
0.0 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 2 |
Hb_003078_060 |
0.2625024234 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 3 |
Hb_000679_120 |
0.2644623707 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 4 |
Hb_003750_070 |
0.2664788493 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005489_030 |
0.2681007755 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 6 |
Hb_000321_080 |
0.2696954742 |
- |
- |
histone H4 [Zea mays] |
| 7 |
Hb_004545_020 |
0.2708226684 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 8 |
Hb_001016_010 |
0.2722655419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004162_230 |
0.2736293212 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 10 |
Hb_005725_200 |
0.2826901646 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 11 |
Hb_000181_190 |
0.2912140796 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 12 |
Hb_032208_020 |
0.3023497761 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 13 |
Hb_000139_490 |
0.3060005243 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 14 |
Hb_002259_200 |
0.3062651078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005679_080 |
0.3073088253 |
- |
- |
PREDICTED: PH-interacting protein-like [Populus euphratica] |
| 16 |
Hb_001300_210 |
0.3147402796 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 17 |
Hb_044662_050 |
0.3173915116 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 18 |
Hb_002411_060 |
0.3191763368 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 19 |
Hb_036790_030 |
0.3211400835 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 20 |
Hb_000739_270 |
0.3235158064 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |