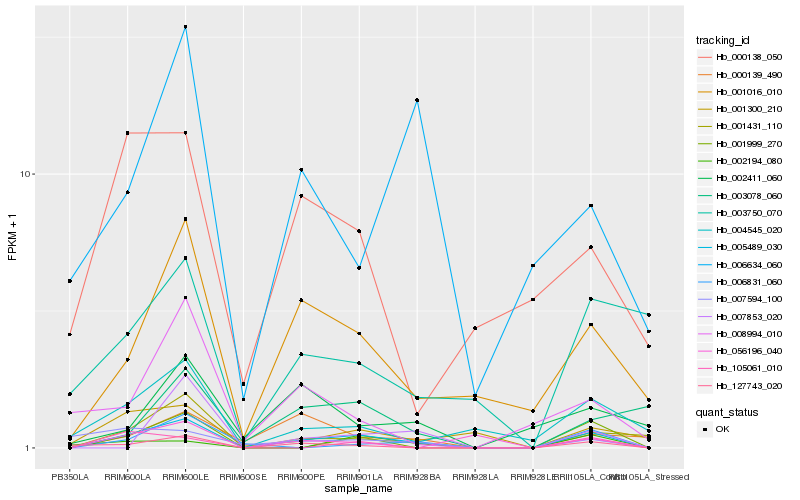
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005489_030 |
0.0 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 2 |
Hb_001431_110 |
0.2681007755 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 3 |
Hb_001016_010 |
0.2836689315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_127743_020 |
0.3002027532 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001999_270 |
0.3022765568 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 6 |
Hb_006831_060 |
0.3241578329 |
- |
- |
hypothetical protein JCGZ_16419 [Jatropha curcas] |
| 7 |
Hb_004545_020 |
0.325287265 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 8 |
Hb_001300_210 |
0.3372778894 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 9 |
Hb_007853_020 |
0.3482333959 |
- |
- |
protein BOLA2 [Jatropha curcas] |
| 10 |
Hb_007594_100 |
0.3498602399 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 11 |
Hb_000139_490 |
0.3515067425 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 12 |
Hb_003078_060 |
0.3531484494 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 13 |
Hb_105061_010 |
0.3541983774 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 14 |
Hb_056196_040 |
0.3605902053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006634_060 |
0.3612458853 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 16 |
Hb_000138_050 |
0.3614523662 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper protein HAT14, putative [Ricinus communis] |
| 17 |
Hb_003750_070 |
0.362361282 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_008994_010 |
0.3647818451 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_002411_060 |
0.365378755 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 20 |
Hb_002194_080 |
0.3663227118 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRD, chloroplastic [Jatropha curcas] |