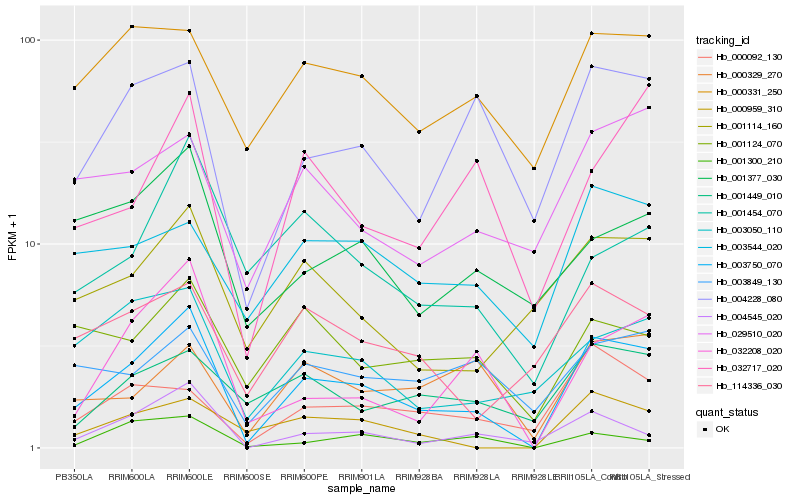
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003750_070 |
0.0 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_032208_020 |
0.2060578309 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 3 |
Hb_004545_020 |
0.2081456259 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 4 |
Hb_004228_080 |
0.2100713375 |
- |
- |
phosphoethanolamine n-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000959_310 |
0.2134491495 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 6 |
Hb_003050_110 |
0.2155029035 |
- |
- |
PREDICTED: prefoldin subunit 6 [Populus euphratica] |
| 7 |
Hb_000092_130 |
0.2247791105 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_000329_270 |
0.2258973355 |
- |
- |
PREDICTED: spindle and kinetochore-associated protein 1 homolog [Jatropha curcas] |
| 9 |
Hb_001114_160 |
0.22620586 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 10 |
Hb_032717_020 |
0.2295695271 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_029510_020 |
0.2295751563 |
- |
- |
hypothetical protein JCGZ_06306 [Jatropha curcas] |
| 12 |
Hb_001454_070 |
0.2308983638 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 13 |
Hb_000331_250 |
0.23100253 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 14 |
Hb_001124_070 |
0.2327438307 |
- |
- |
Serine/threonine-protein kinase ULK4 [Gossypium arboreum] |
| 15 |
Hb_003544_020 |
0.232830621 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 16 |
Hb_114336_030 |
0.2329859749 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 17 |
Hb_001300_210 |
0.2331229529 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 18 |
Hb_003849_130 |
0.2332388561 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 19 |
Hb_001377_030 |
0.2335725401 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 20 |
Hb_001449_010 |
0.2339799697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |