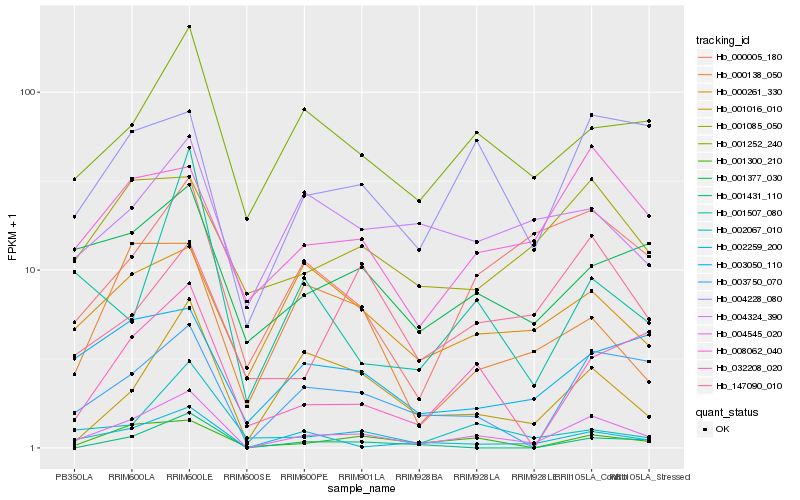
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004545_020 |
0.0 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 2 |
Hb_001300_210 |
0.2051257642 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 3 |
Hb_002259_200 |
0.2079185459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003750_070 |
0.2081456259 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001252_240 |
0.217612919 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 6 |
Hb_001016_010 |
0.2280836642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008062_040 |
0.237093912 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 8 |
Hb_032208_020 |
0.2378025181 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 9 |
Hb_000005_180 |
0.2385377824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004228_080 |
0.2386796686 |
- |
- |
phosphoethanolamine n-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_002067_010 |
0.2427123915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001377_030 |
0.2430037429 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 13 |
Hb_000138_050 |
0.248060047 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper protein HAT14, putative [Ricinus communis] |
| 14 |
Hb_001085_050 |
0.2484867483 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 15 |
Hb_003050_110 |
0.2524430422 |
- |
- |
PREDICTED: prefoldin subunit 6 [Populus euphratica] |
| 16 |
Hb_000261_330 |
0.2565579243 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Amborella trichopoda] |
| 17 |
Hb_001507_080 |
0.2619333266 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 18 |
Hb_004324_390 |
0.266574418 |
- |
- |
PREDICTED: plastidial pyruvate kinase 2 [Jatropha curcas] |
| 19 |
Hb_001431_110 |
0.2708226684 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 20 |
Hb_147090_010 |
0.2713462431 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Malus domestica] |