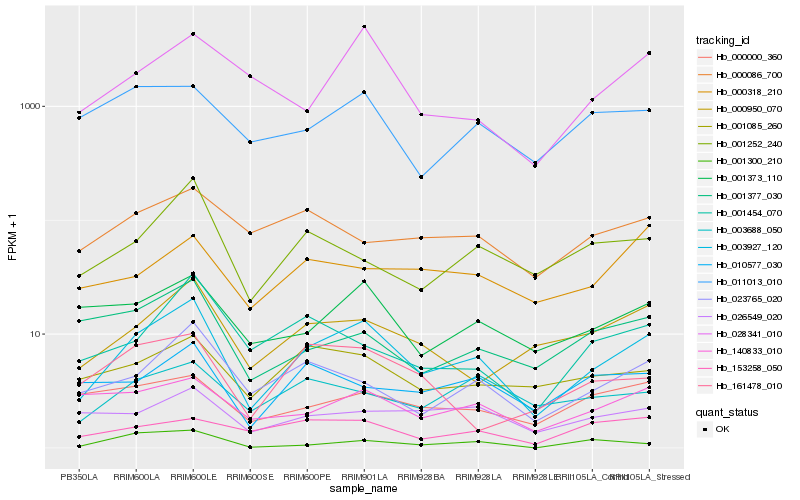
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003927_120 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 2 |
Hb_023765_020 |
0.184647079 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 3 |
Hb_000950_070 |
0.1886571361 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001373_110 |
0.1938033169 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 5 |
Hb_001252_240 |
0.1987510844 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 6 |
Hb_001085_260 |
0.2003690783 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 7 |
Hb_140833_010 |
0.2031616957 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 8 |
Hb_028341_010 |
0.2063449254 |
- |
- |
hypothetical protein CICLE_v10026841mg [Citrus clementina] |
| 9 |
Hb_003688_050 |
0.2077603587 |
- |
- |
PREDICTED: uncharacterized protein LOC105640616 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001454_070 |
0.2090403733 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 11 |
Hb_026549_020 |
0.2100352278 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001377_030 |
0.2134350616 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 13 |
Hb_010577_030 |
0.2145134446 |
- |
- |
PREDICTED: putative cyclin-B3-1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000000_360 |
0.2145535569 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 15 |
Hb_011013_010 |
0.2157093792 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 16 |
Hb_153258_050 |
0.2158637559 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 17 |
Hb_161478_010 |
0.2189022928 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000318_210 |
0.2231154091 |
- |
- |
PREDICTED: 40S ribosomal protein S12-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000086_700 |
0.225053458 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 20 |
Hb_001300_210 |
0.2258616638 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |