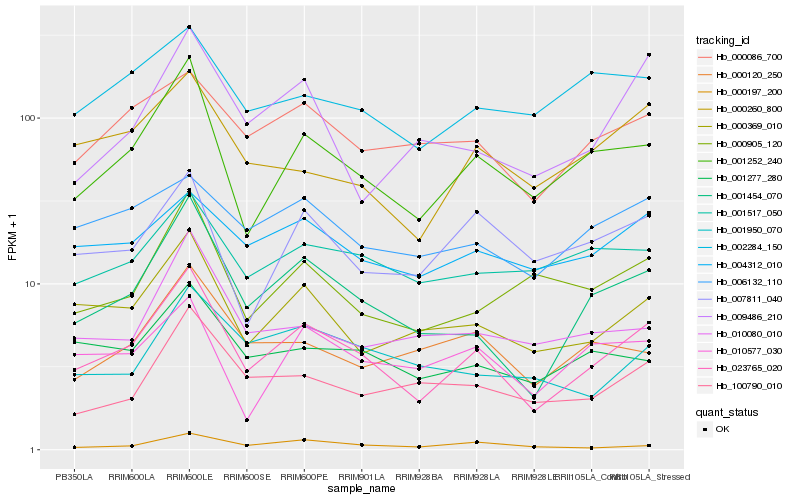
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023765_020 |
0.0 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 2 |
Hb_001454_070 |
0.0968750904 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 3 |
Hb_001252_240 |
0.1131635026 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 4 |
Hb_000369_010 |
0.1340610792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000086_700 |
0.1549317002 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 6 |
Hb_000260_800 |
0.1561231545 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 7 |
Hb_001277_280 |
0.1596396786 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 8 |
Hb_100790_010 |
0.1609170753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000905_120 |
0.161954477 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 10 |
Hb_000197_200 |
0.1632183794 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 11 |
Hb_000120_250 |
0.1664340486 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009486_210 |
0.1673663006 |
- |
- |
- |
| 13 |
Hb_006132_110 |
0.1675540245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010577_030 |
0.1677605588 |
- |
- |
PREDICTED: putative cyclin-B3-1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_010080_010 |
0.1678976406 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 16 |
Hb_007811_040 |
0.1684146068 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 isoform X1 [Gossypium raimondii] |
| 17 |
Hb_004312_010 |
0.1686394191 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001950_070 |
0.1698478057 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 19 |
Hb_001517_050 |
0.1712866153 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002284_150 |
0.1722210903 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |