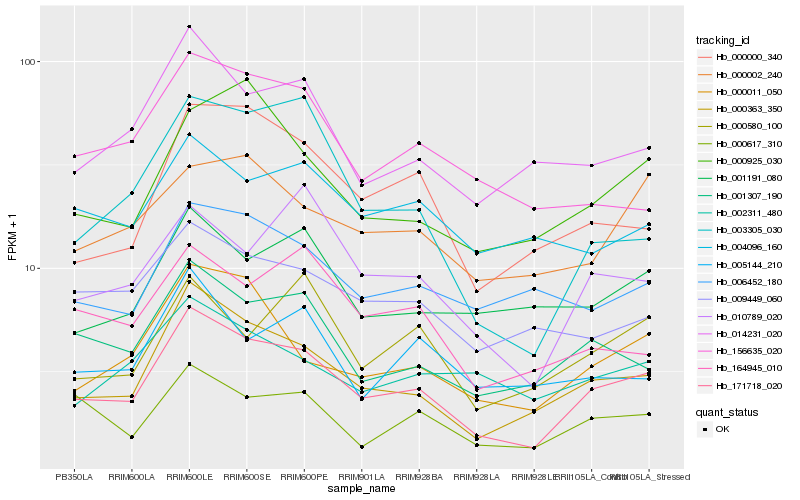
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171718_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 2 |
Hb_000363_350 |
0.0852306498 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |
| 3 |
Hb_001307_190 |
0.1233184564 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000011_050 |
0.1252856186 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 5 |
Hb_014231_020 |
0.1334098882 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 6 |
Hb_000000_340 |
0.1351719871 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 29-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000617_310 |
0.1380259087 |
- |
- |
PREDICTED: cell cycle checkpoint control protein RAD9A [Jatropha curcas] |
| 8 |
Hb_000925_030 |
0.1414643519 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 9 |
Hb_010789_020 |
0.143554273 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 10 |
Hb_009449_060 |
0.1457012845 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 11 |
Hb_006452_180 |
0.1474865808 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 12 |
Hb_001191_080 |
0.1490903586 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000580_100 |
0.1496287719 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000002_240 |
0.1499029075 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 15 |
Hb_164945_010 |
0.1509408597 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 16 |
Hb_002311_480 |
0.1520932331 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003305_030 |
0.1523374677 |
- |
- |
- |
| 18 |
Hb_156635_020 |
0.1524408735 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 19 |
Hb_004096_160 |
0.1535722091 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 20 |
Hb_005144_210 |
0.1536347299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |