| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
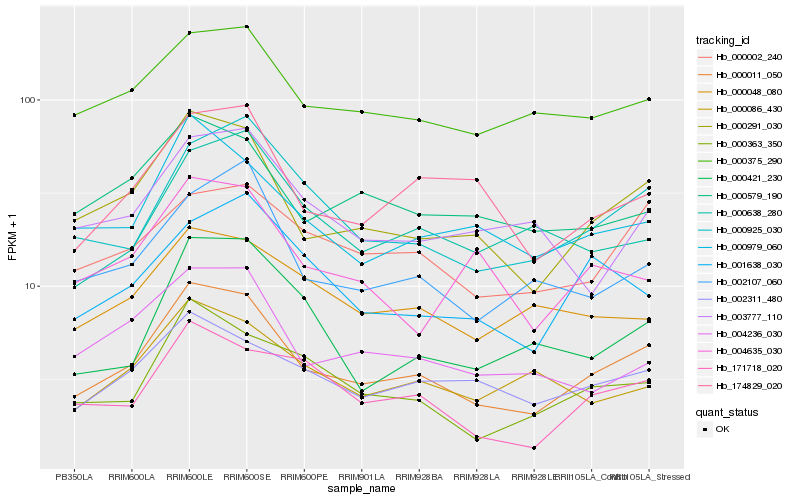
Hb_000011_050 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 2 |
Hb_000291_030 |
0.0727106767 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 3 |
Hb_000925_030 |
0.117031036 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_000363_350 |
0.1181628505 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |
| 5 |
Hb_000979_060 |
0.1199670721 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 6 |
Hb_174829_020 |
0.1228156108 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 7 |
Hb_002311_480 |
0.1243315168 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_171718_020 |
0.1252856186 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 9 |
Hb_000375_290 |
0.1253786349 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_002107_060 |
0.1256614576 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 11 |
Hb_004236_030 |
0.1284087955 |
- |
- |
unknown [Lotus japonicus] |
| 12 |
Hb_000579_190 |
0.1324328441 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 13 |
Hb_000002_240 |
0.1344938423 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 14 |
Hb_000086_430 |
0.1358988652 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 15 |
Hb_000048_080 |
0.1380065554 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000421_230 |
0.138393273 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 17 |
Hb_001638_030 |
0.1398734789 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 18 |
Hb_000638_280 |
0.1420565087 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 19 |
Hb_004635_030 |
0.1439744752 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 20 |
Hb_003777_110 |
0.1444404496 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 8 [Jatropha curcas] |