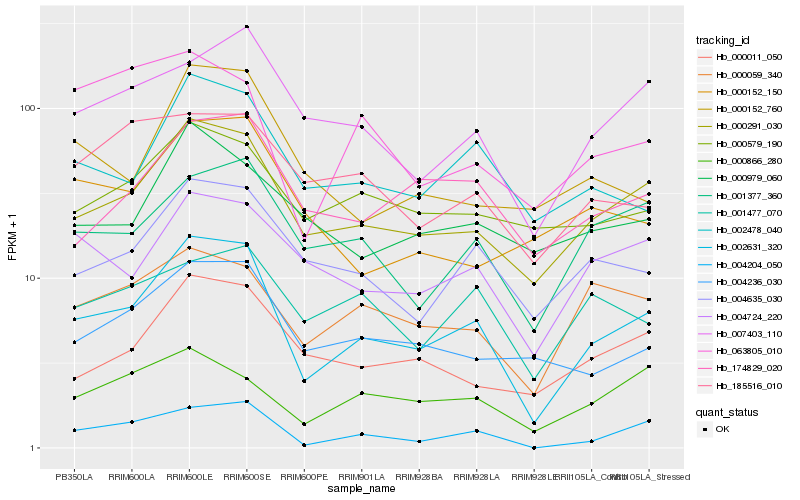
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_320 |
0.0 |
- |
- |
hypothetical protein JCGZ_22628 [Jatropha curcas] |
| 2 |
Hb_000291_030 |
0.1073397922 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 3 |
Hb_004204_050 |
0.1272116929 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_004635_030 |
0.1530427582 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 5 |
Hb_002478_040 |
0.1546435675 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 6 |
Hb_001377_360 |
0.1580180817 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 7 |
Hb_000011_050 |
0.1602933877 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 8 |
Hb_007403_110 |
0.1604502422 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 9 |
Hb_004236_030 |
0.1613112772 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_004724_220 |
0.165177155 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 11 |
Hb_174829_020 |
0.1654506188 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 12 |
Hb_000059_340 |
0.1710776916 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 13 |
Hb_000579_190 |
0.1750634156 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 14 |
Hb_000152_760 |
0.1792041464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000866_280 |
0.1801845298 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 16 |
Hb_000979_060 |
0.1803103107 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 17 |
Hb_185516_010 |
0.1850202146 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 18 |
Hb_001477_070 |
0.1862491792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000152_150 |
0.1899825274 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 20 |
Hb_063805_010 |
0.1912466005 |
- |
- |
- |