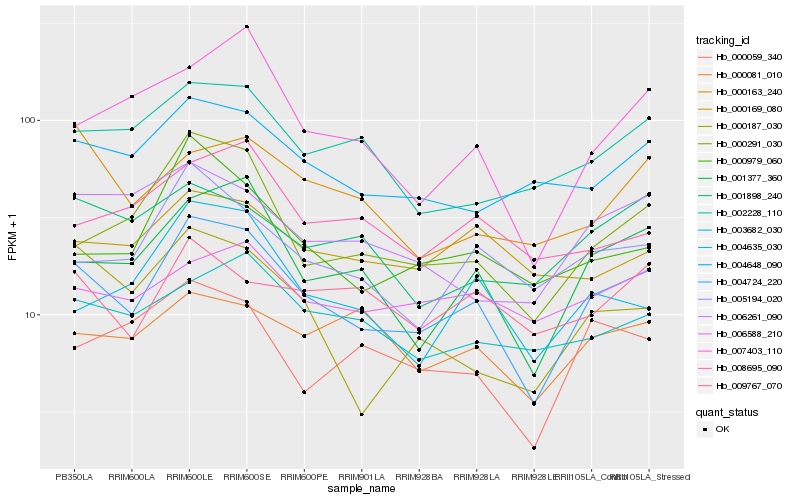
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_220 |
0.0 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 2 |
Hb_001377_360 |
0.108127704 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 3 |
Hb_000291_030 |
0.1218844536 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 4 |
Hb_005194_020 |
0.1298882188 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004648_090 |
0.1343782426 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 6 |
Hb_000979_060 |
0.1357643217 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 7 |
Hb_004635_030 |
0.1370002969 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 8 |
Hb_002228_110 |
0.1403695705 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 9 |
Hb_007403_110 |
0.1405414481 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 10 |
Hb_003682_030 |
0.1418045284 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 11 |
Hb_006261_090 |
0.1423288916 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 12 |
Hb_000187_030 |
0.1451750315 |
- |
- |
PREDICTED: cyclin-D4-1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000059_340 |
0.145609685 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 14 |
Hb_006588_210 |
0.1478540246 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 15 |
Hb_000169_080 |
0.1481350768 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 16 |
Hb_000081_010 |
0.1498085262 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 17 |
Hb_001898_240 |
0.1502344498 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 18 |
Hb_009767_070 |
0.1503086926 |
- |
- |
PREDICTED: uncharacterized protein LOC105631980 [Jatropha curcas] |
| 19 |
Hb_008695_090 |
0.1509010213 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 20 |
Hb_000163_240 |
0.1526450304 |
- |
- |
PREDICTED: protein OPI10 homolog [Jatropha curcas] |