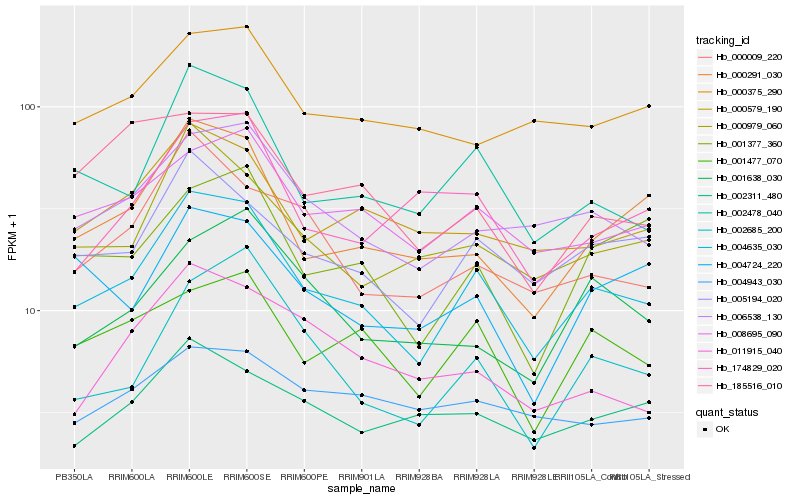
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004635_030 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 2 |
Hb_002478_040 |
0.0976125698 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 3 |
Hb_005194_020 |
0.1020751208 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_006538_130 |
0.1042467463 |
- |
- |
Uncharacterized protein TCM_009867 [Theobroma cacao] |
| 5 |
Hb_008695_090 |
0.1118812677 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 6 |
Hb_000291_030 |
0.1152178044 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 7 |
Hb_001477_070 |
0.1163713026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000979_060 |
0.1176174947 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 9 |
Hb_000579_190 |
0.1239051724 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 10 |
Hb_001377_360 |
0.1240520153 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 11 |
Hb_000009_220 |
0.1301473299 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_002685_200 |
0.1328936344 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 13 |
Hb_002311_480 |
0.1350168012 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000375_290 |
0.1362665726 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_174829_020 |
0.1365751957 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 16 |
Hb_004724_220 |
0.1370002969 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 17 |
Hb_001638_030 |
0.1376946363 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 18 |
Hb_185516_010 |
0.1427121346 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 19 |
Hb_011915_040 |
0.1436035398 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 20 |
Hb_004943_030 |
0.1439586095 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |