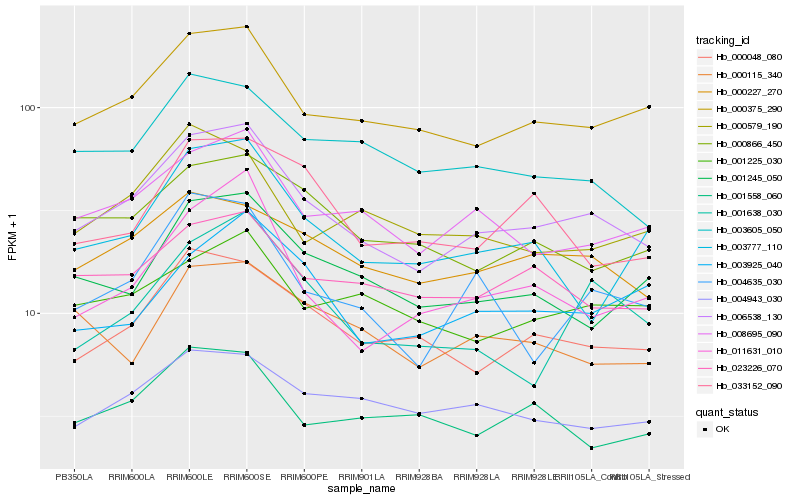
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_130 |
0.0 |
- |
- |
Uncharacterized protein TCM_009867 [Theobroma cacao] |
| 2 |
Hb_000375_290 |
0.0795646394 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_008695_090 |
0.0903251401 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 4 |
Hb_000048_080 |
0.0918492311 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000227_270 |
0.0941885287 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_023226_070 |
0.1027520068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004635_030 |
0.1042467463 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 8 |
Hb_001245_050 |
0.104459802 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 9 |
Hb_003605_050 |
0.1051374646 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000866_450 |
0.1066739315 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_004943_030 |
0.1070577636 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 12 |
Hb_011631_010 |
0.1097742528 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 13 |
Hb_001225_030 |
0.1119604502 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 14 |
Hb_001638_030 |
0.1137377222 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 15 |
Hb_001558_060 |
0.1155974044 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000579_190 |
0.1160701255 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 17 |
Hb_003777_110 |
0.1161323018 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 8 [Jatropha curcas] |
| 18 |
Hb_003925_040 |
0.117792745 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 19 |
Hb_033152_090 |
0.1182107545 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 20 |
Hb_000115_340 |
0.1190056188 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |