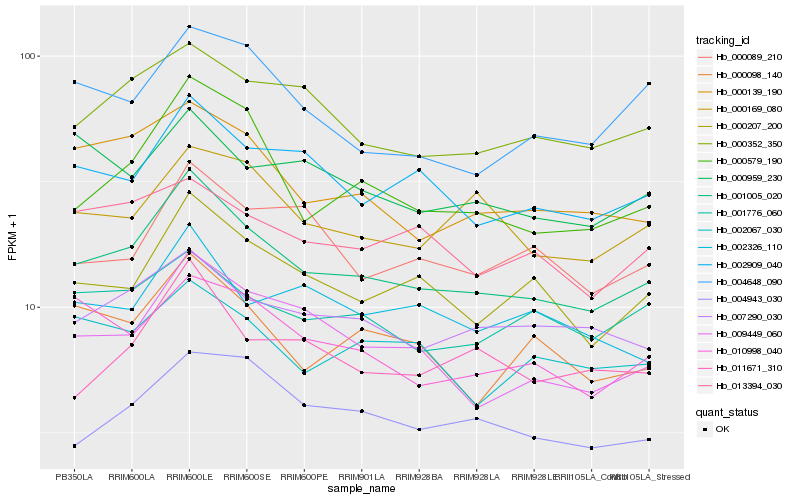
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001005_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 2 |
Hb_000207_200 |
0.0725934945 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000579_190 |
0.0729908645 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 4 |
Hb_002909_040 |
0.0753407501 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 5 |
Hb_000352_350 |
0.0772576509 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 6 |
Hb_009449_060 |
0.0793717257 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 7 |
Hb_007290_030 |
0.0796787313 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 8 |
Hb_000139_190 |
0.0800580311 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000169_080 |
0.0871857194 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 10 |
Hb_011671_310 |
0.0876427582 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 11 |
Hb_004943_030 |
0.0879936634 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 12 |
Hb_010998_040 |
0.0886223392 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 13 |
Hb_001776_060 |
0.0895614897 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 14 |
Hb_000098_140 |
0.0904310224 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 15 |
Hb_000089_210 |
0.0914725245 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_002067_030 |
0.0924420037 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_013394_030 |
0.0928950535 |
- |
- |
PREDICTED: exocyst complex component SEC6 [Vitis vinifera] |
| 18 |
Hb_000959_230 |
0.0929005843 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 19 |
Hb_004648_090 |
0.0943358357 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 20 |
Hb_002326_110 |
0.0951892465 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |