| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
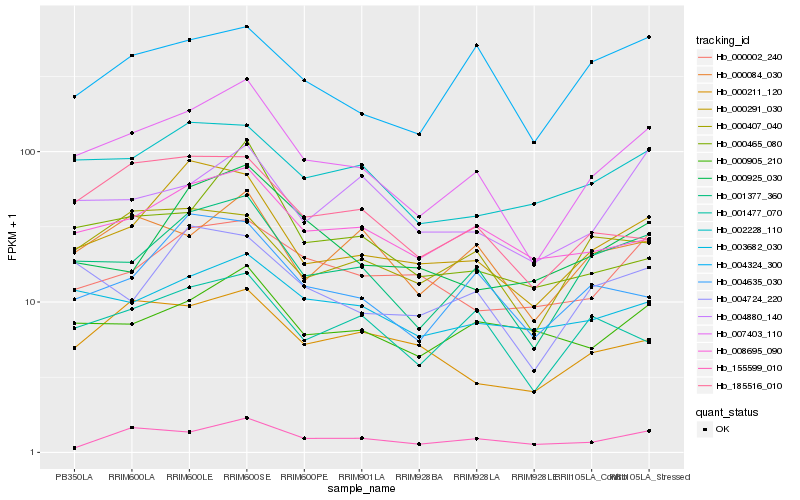
Hb_007403_110 |
0.0 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 2 |
Hb_001377_360 |
0.0801131662 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 3 |
Hb_000291_030 |
0.1289027632 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 4 |
Hb_008695_090 |
0.1357111596 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 5 |
Hb_002228_110 |
0.1378114499 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 6 |
Hb_000084_030 |
0.1392726482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004724_220 |
0.1405414481 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 8 |
Hb_003682_030 |
0.1415616009 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 9 |
Hb_185516_010 |
0.1465408713 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 10 |
Hb_001477_070 |
0.1468054742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000211_120 |
0.1485620035 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 12 |
Hb_004635_030 |
0.1489243941 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 13 |
Hb_000905_210 |
0.1491027847 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 14 |
Hb_000465_080 |
0.1507122556 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 15 |
Hb_155599_010 |
0.1517143375 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_004880_140 |
0.1537978901 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 17 |
Hb_000002_240 |
0.1545293003 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 18 |
Hb_004324_300 |
0.1546566789 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 19 |
Hb_000407_040 |
0.1551496857 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000925_030 |
0.1562983365 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |