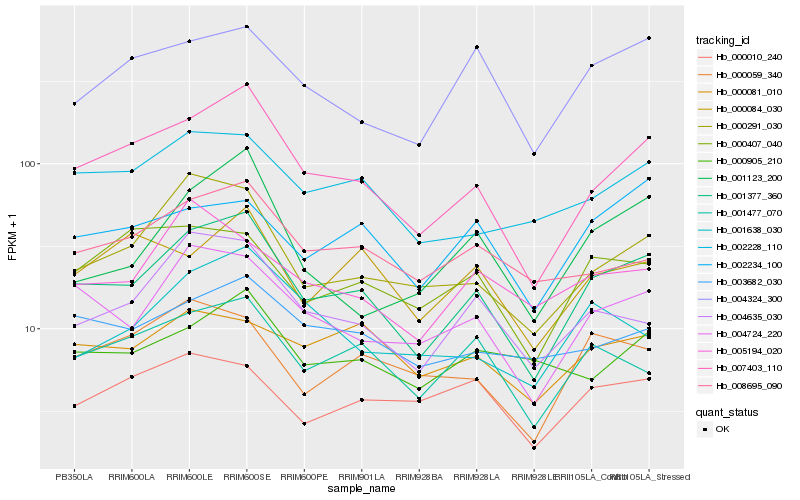
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_360 |
0.0 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 2 |
Hb_007403_110 |
0.0801131662 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 3 |
Hb_004724_220 |
0.108127704 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 4 |
Hb_000291_030 |
0.1194817253 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 5 |
Hb_001477_070 |
0.123858925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004635_030 |
0.1240520153 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 7 |
Hb_002228_110 |
0.1276072904 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 8 |
Hb_004324_300 |
0.1329940623 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 9 |
Hb_008695_090 |
0.1362360735 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 10 |
Hb_000059_340 |
0.1398089406 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 11 |
Hb_003682_030 |
0.1398779582 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 12 |
Hb_001123_200 |
0.1408737323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000407_040 |
0.1439950302 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_002234_100 |
0.1455340601 |
- |
- |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 15 |
Hb_000084_030 |
0.1458815657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000905_210 |
0.1471077874 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 17 |
Hb_000081_010 |
0.1488331638 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 18 |
Hb_005194_020 |
0.1497353149 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001638_030 |
0.1521832321 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 20 |
Hb_000010_240 |
0.1529134756 |
- |
- |
Protein bem46, putative [Ricinus communis] |