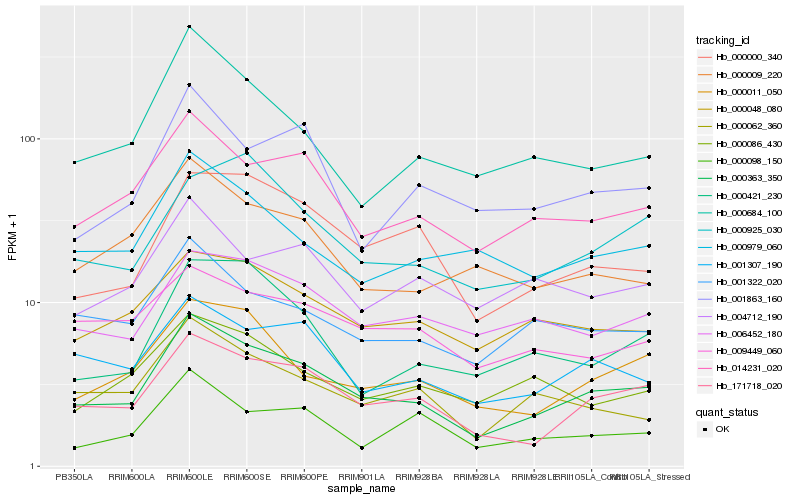
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_350 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |
| 2 |
Hb_171718_020 |
0.0852306498 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 3 |
Hb_014231_020 |
0.107639975 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 4 |
Hb_000011_050 |
0.1181628505 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 5 |
Hb_001307_190 |
0.1183501665 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000000_340 |
0.1246287036 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 29-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000062_360 |
0.1253890869 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 8 |
Hb_001322_020 |
0.1269579495 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 9 |
Hb_000048_080 |
0.1280227586 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006452_180 |
0.1326736112 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 11 |
Hb_000979_060 |
0.1354223248 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 12 |
Hb_000684_100 |
0.1358734451 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 13 |
Hb_000009_220 |
0.1378410866 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000925_030 |
0.1386389132 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_000421_230 |
0.1401863325 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 16 |
Hb_001863_160 |
0.1407992621 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 17 |
Hb_000086_430 |
0.1429937876 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 18 |
Hb_009449_060 |
0.1432584632 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 19 |
Hb_004712_190 |
0.1437715003 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 20 |
Hb_000098_150 |
0.1438630306 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |