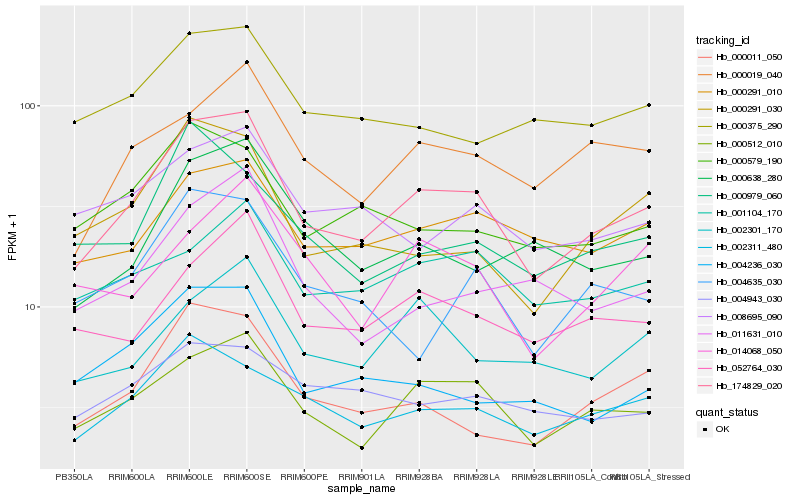
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174829_020 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 2 |
Hb_000512_010 |
0.0850176134 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 3 |
Hb_002311_480 |
0.1173107252 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000291_010 |
0.1189560504 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 5 |
Hb_052764_030 |
0.119812655 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 6 |
Hb_000019_040 |
0.1205563935 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000011_050 |
0.1228156108 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 8 |
Hb_000291_030 |
0.1270826264 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 9 |
Hb_000638_280 |
0.1294883469 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 10 |
Hb_000579_190 |
0.12959366 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 11 |
Hb_002301_170 |
0.1307270113 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |
| 12 |
Hb_004236_030 |
0.1323600512 |
- |
- |
unknown [Lotus japonicus] |
| 13 |
Hb_000375_290 |
0.1324117707 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_008695_090 |
0.1330697935 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 15 |
Hb_001104_170 |
0.1348634365 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 16 |
Hb_011631_010 |
0.1348770971 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 17 |
Hb_004635_030 |
0.1365751957 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 18 |
Hb_014068_050 |
0.1378432321 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 19 |
Hb_000979_060 |
0.1382609363 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 20 |
Hb_004943_030 |
0.139220916 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |