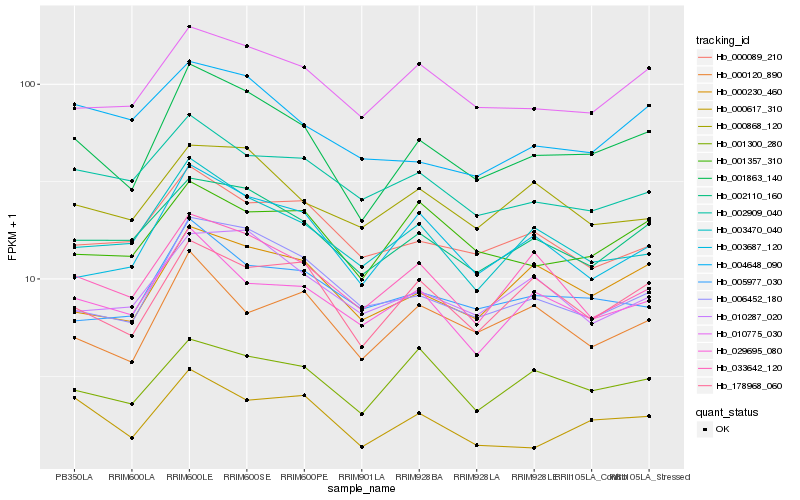
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_140 |
0.0 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6b-1 [Jatropha curcas] |
| 2 |
Hb_002110_160 |
0.0854142827 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_006452_180 |
0.094034481 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 4 |
Hb_178968_060 |
0.0940965438 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 5 |
Hb_000230_460 |
0.0963449533 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 6 |
Hb_010775_030 |
0.0969713735 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 7 |
Hb_003687_120 |
0.0993781199 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 8 |
Hb_003470_040 |
0.099870123 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_029695_080 |
0.1016703379 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 10 |
Hb_033642_120 |
0.1051758649 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 11 |
Hb_005977_030 |
0.1074552035 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 12 |
Hb_010287_020 |
0.1092045635 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 13 |
Hb_001300_280 |
0.1092157175 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 14 |
Hb_000868_120 |
0.1101802837 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 15 |
Hb_000089_210 |
0.1111287326 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_001357_310 |
0.1119662983 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000120_890 |
0.1139933931 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 18 |
Hb_002909_040 |
0.1143305228 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 19 |
Hb_004648_090 |
0.1145909724 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 20 |
Hb_000617_310 |
0.1146473217 |
- |
- |
PREDICTED: cell cycle checkpoint control protein RAD9A [Jatropha curcas] |