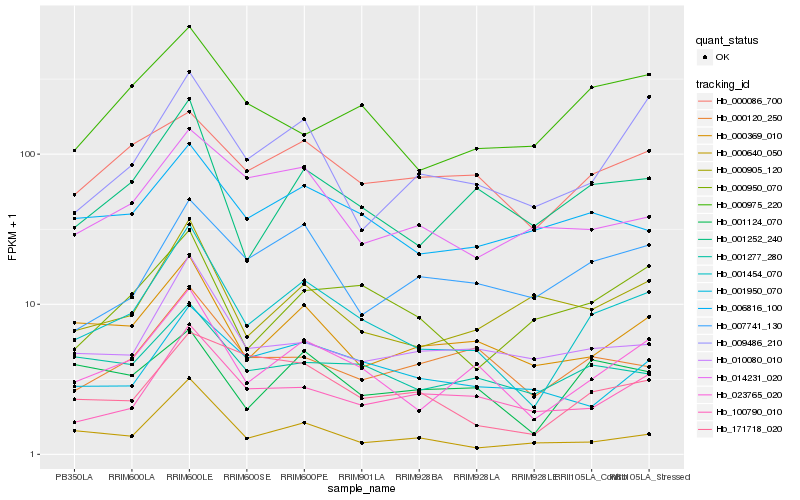
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_070 |
0.0 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 2 |
Hb_023765_020 |
0.0968750904 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 3 |
Hb_001252_240 |
0.1476521624 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 4 |
Hb_000369_010 |
0.1543283496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_100790_010 |
0.1632277438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000086_700 |
0.1658628914 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 7 |
Hb_171718_020 |
0.1662437192 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 8 |
Hb_001277_280 |
0.166382187 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 9 |
Hb_009486_210 |
0.1667409563 |
- |
- |
- |
| 10 |
Hb_010080_010 |
0.1677339774 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 11 |
Hb_006816_100 |
0.1691891262 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 12 |
Hb_000640_050 |
0.1698527231 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 13 |
Hb_001950_070 |
0.1713754148 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 14 |
Hb_000120_250 |
0.1721458457 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001124_070 |
0.1728042534 |
- |
- |
Serine/threonine-protein kinase ULK4 [Gossypium arboreum] |
| 16 |
Hb_000975_220 |
0.1749279293 |
- |
- |
Histone H2B [Medicago truncatula] |
| 17 |
Hb_000950_070 |
0.1754662404 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000905_120 |
0.176077104 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 19 |
Hb_007741_130 |
0.1768652632 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 20 |
Hb_014231_020 |
0.1786741763 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |