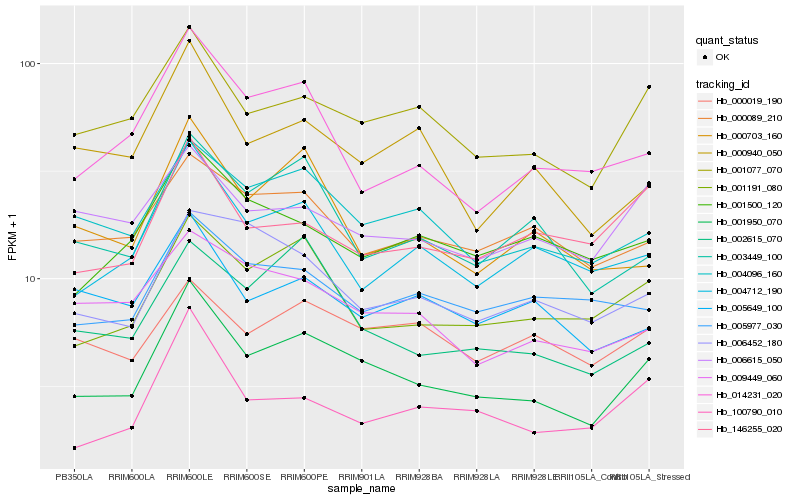
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001950_070 |
0.0 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 2 |
Hb_001077_070 |
0.0942888373 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 3 |
Hb_001191_080 |
0.1077068969 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_004096_160 |
0.1085834415 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 5 |
Hb_004712_190 |
0.1169151679 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 6 |
Hb_003449_100 |
0.121611892 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 7 |
Hb_100790_010 |
0.1240959337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000940_050 |
0.1249087254 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 9 |
Hb_002615_070 |
0.1251259839 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 10 |
Hb_014231_020 |
0.1254721745 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 11 |
Hb_000703_160 |
0.1265077234 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_009449_060 |
0.1270235861 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 13 |
Hb_001500_120 |
0.1280913476 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_000089_210 |
0.128146299 |
- |
- |
unknown [Medicago truncatula] |
| 15 |
Hb_006452_180 |
0.1311857275 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 16 |
Hb_005977_030 |
0.1316248306 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 17 |
Hb_006615_050 |
0.1318276015 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 18 |
Hb_146255_020 |
0.1318710179 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 19 |
Hb_005649_100 |
0.1321131399 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000019_190 |
0.1327452501 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |