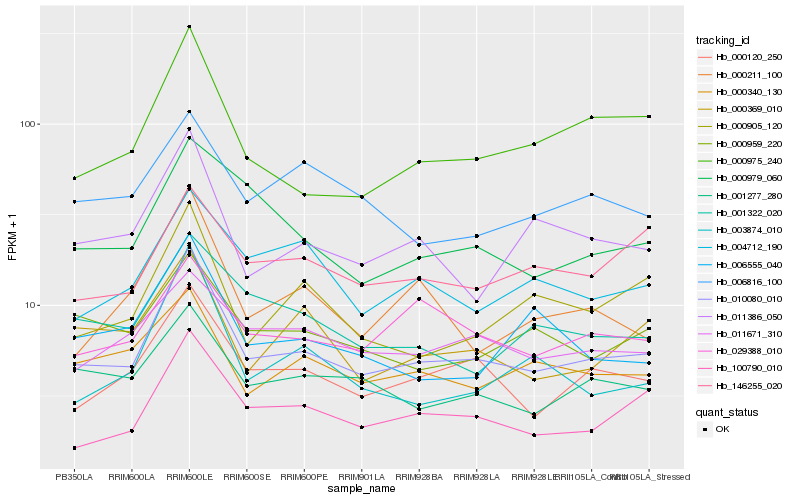
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010080_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 2 |
Hb_000120_250 |
0.0993123024 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 3 |
Hb_100790_010 |
0.1053969769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000975_240 |
0.1125456563 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 5 |
Hb_011386_050 |
0.1156830004 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001277_280 |
0.1173491197 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 7 |
Hb_000340_130 |
0.1247918501 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000905_120 |
0.1255002072 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 9 |
Hb_000979_060 |
0.1258036467 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 10 |
Hb_001322_020 |
0.12588366 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 11 |
Hb_000211_100 |
0.1271464284 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 12 |
Hb_000369_010 |
0.1273633146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003874_010 |
0.1279315447 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_029388_010 |
0.1283534635 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 15 |
Hb_000959_220 |
0.1287731526 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_004712_190 |
0.1302291853 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 17 |
Hb_011671_310 |
0.1309407999 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 18 |
Hb_146255_020 |
0.1333212065 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 19 |
Hb_006555_040 |
0.1343514894 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 20 |
Hb_006816_100 |
0.1360073348 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |