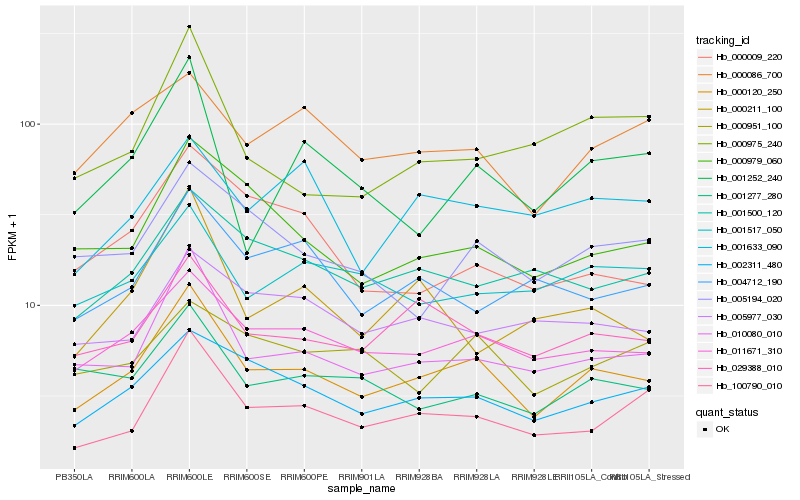
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630152 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011671_310 |
0.0983536899 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 3 |
Hb_010080_010 |
0.0993123024 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 4 |
Hb_029388_010 |
0.1136843763 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |
| 5 |
Hb_002311_480 |
0.1157157839 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_100790_010 |
0.1180631445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000979_060 |
0.1275328662 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 8 |
Hb_001500_120 |
0.1317712227 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_001277_280 |
0.1358701258 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 10 |
Hb_005194_020 |
0.1370110994 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000086_700 |
0.1372475416 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 12 |
Hb_001517_050 |
0.1391319034 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005977_030 |
0.1436344123 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 14 |
Hb_004712_190 |
0.1442558836 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 15 |
Hb_001633_090 |
0.1490324367 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001252_240 |
0.1494777753 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 17 |
Hb_000951_100 |
0.150773209 |
- |
- |
PREDICTED: uncharacterized protein LOC103436565 isoform X1 [Malus domestica] |
| 18 |
Hb_000009_220 |
0.1510277786 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000975_240 |
0.151220347 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 20 |
Hb_000211_100 |
0.1512250047 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |