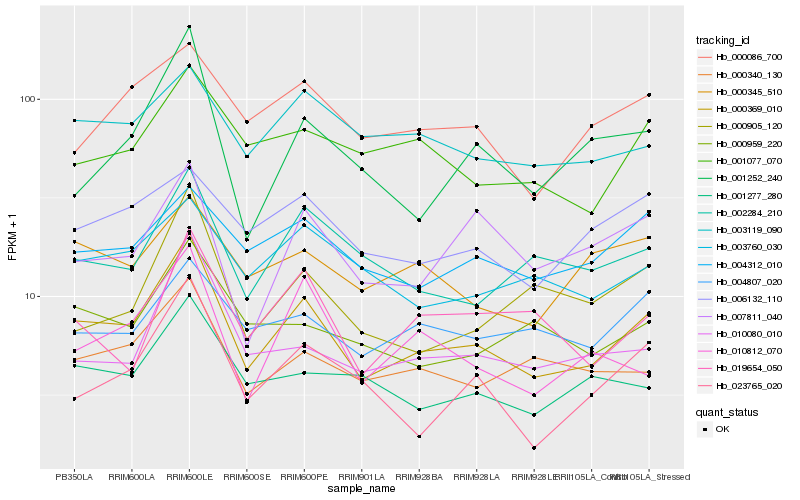
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000369_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000340_130 |
0.1233631399 |
- |
- |
PREDICTED: uncharacterized protein LOC105650033 isoform X1 [Jatropha curcas] |
| 3 |
Hb_010080_010 |
0.1273633146 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 4 |
Hb_002284_210 |
0.1308722568 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 5 |
Hb_001277_280 |
0.1324442965 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 6 |
Hb_023765_020 |
0.1340610792 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 7 |
Hb_000959_220 |
0.1352734346 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001077_070 |
0.1364101145 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 9 |
Hb_003119_090 |
0.1367146969 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 10 |
Hb_010812_070 |
0.1374743381 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 11 |
Hb_003760_030 |
0.1376846376 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 12 |
Hb_001252_240 |
0.1377908775 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 13 |
Hb_004312_010 |
0.1380891709 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000086_700 |
0.1384108339 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 15 |
Hb_000905_120 |
0.1385428922 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 16 |
Hb_004807_020 |
0.1395580094 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 17 |
Hb_007811_040 |
0.1405917722 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 isoform X1 [Gossypium raimondii] |
| 18 |
Hb_019654_050 |
0.1407203438 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 19 |
Hb_006132_110 |
0.1409691507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000345_510 |
0.1410029468 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |