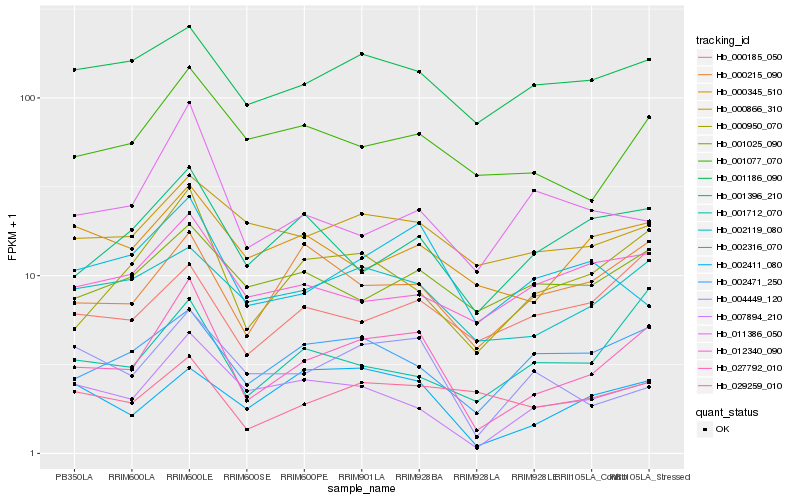
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027792_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000950_070 |
0.1396621138 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007894_210 |
0.1704665295 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 4 |
Hb_002471_250 |
0.1731973797 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |
| 5 |
Hb_002119_080 |
0.1740388799 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000215_090 |
0.1791572761 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_001077_070 |
0.1801107455 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 8 |
Hb_001396_210 |
0.1846840667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002411_080 |
0.1859977534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004449_120 |
0.1887213341 |
- |
- |
PREDICTED: uncharacterized protein LOC105631291 [Jatropha curcas] |
| 11 |
Hb_001712_070 |
0.1923178722 |
- |
- |
PREDICTED: HD domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_001186_090 |
0.1928745803 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 13 |
Hb_002316_070 |
0.1935929201 |
- |
- |
PREDICTED: 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000345_510 |
0.1955698723 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 15 |
Hb_029259_010 |
0.1962844938 |
- |
- |
hypothetical protein JCGZ_20186 [Jatropha curcas] |
| 16 |
Hb_000185_050 |
0.1963668024 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 17 |
Hb_011386_050 |
0.1964565925 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000866_310 |
0.1973755951 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 19 |
Hb_012340_090 |
0.1978850806 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 20 |
Hb_001025_090 |
0.1989946015 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |