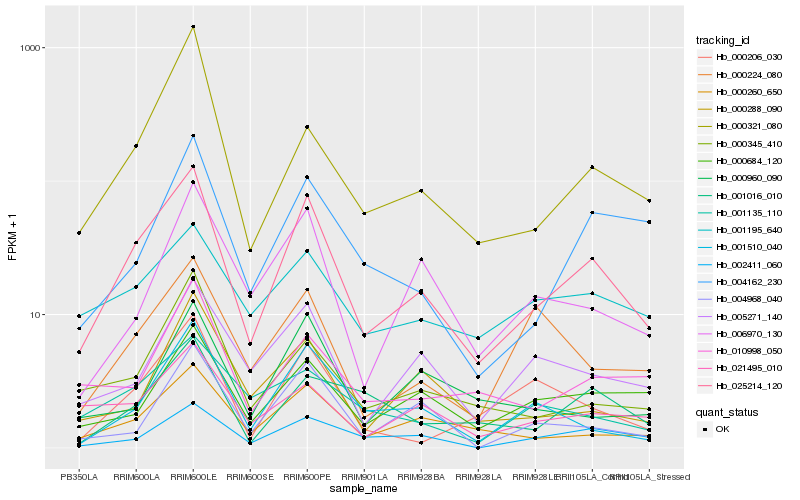
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025214_120 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 2 |
Hb_000260_650 |
0.1660810713 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 3 |
Hb_001510_040 |
0.1919159628 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein [Jatropha curcas] |
| 4 |
Hb_005271_140 |
0.1968938947 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 5 |
Hb_002411_060 |
0.1974305383 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 6 |
Hb_004162_230 |
0.198513075 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 7 |
Hb_000684_120 |
0.1994022283 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 8 |
Hb_006970_130 |
0.1999679941 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 9 |
Hb_000206_030 |
0.2011272445 |
- |
- |
PREDICTED: uncharacterized protein LOC105639682 [Jatropha curcas] |
| 10 |
Hb_000345_410 |
0.2031394072 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 11 |
Hb_000321_080 |
0.2038003001 |
- |
- |
histone H4 [Zea mays] |
| 12 |
Hb_021495_010 |
0.2048166005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004968_040 |
0.2062046866 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 14 |
Hb_010998_050 |
0.2080558272 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000288_090 |
0.2082839953 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000960_090 |
0.2132051268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001135_110 |
0.2134579285 |
- |
- |
- |
| 18 |
Hb_000224_080 |
0.2138385341 |
- |
- |
hypothetical protein RCOM_0510880 [Ricinus communis] |
| 19 |
Hb_001016_010 |
0.2162017469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001195_640 |
0.2168441811 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |