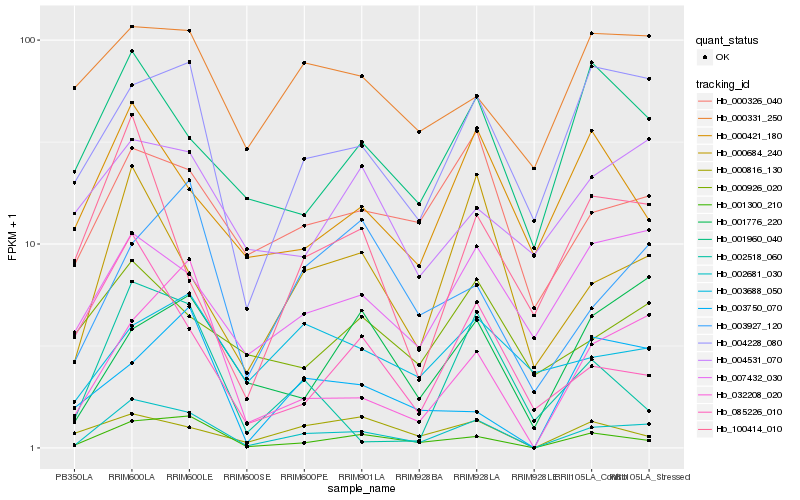
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002681_030 |
0.0 |
- |
- |
PREDICTED: fatty-acid-binding protein 1-like [Jatropha curcas] |
| 2 |
Hb_001300_210 |
0.1695394408 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 3 |
Hb_000684_240 |
0.2093875798 |
- |
- |
hypothetical protein RCOM_0819620 [Ricinus communis] |
| 4 |
Hb_004228_080 |
0.2199739733 |
- |
- |
phosphoethanolamine n-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_032208_020 |
0.2241334823 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 6 |
Hb_000326_040 |
0.2350477381 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 7 |
Hb_007432_030 |
0.2431846234 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 8 |
Hb_001960_040 |
0.2540235582 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 9 |
Hb_000816_130 |
0.2554291012 |
- |
- |
PREDICTED: uncharacterized protein LOC105649213 [Jatropha curcas] |
| 10 |
Hb_003927_120 |
0.255682374 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 11 |
Hb_001776_220 |
0.2596388231 |
- |
- |
- |
| 12 |
Hb_100414_010 |
0.2623737026 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl pyrophosphate synthase [Hevea brasiliensis] |
| 13 |
Hb_000421_180 |
0.2631181446 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000926_020 |
0.2659228448 |
- |
- |
- |
| 15 |
Hb_002518_060 |
0.2685478287 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 16 |
Hb_000331_250 |
0.2702134853 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 17 |
Hb_085226_010 |
0.2729933918 |
- |
- |
hypothetical protein POPTR_0002s03930g [Populus trichocarpa] |
| 18 |
Hb_003688_050 |
0.273869577 |
- |
- |
PREDICTED: uncharacterized protein LOC105640616 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003750_070 |
0.2740499851 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004531_070 |
0.2757300877 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX19-like [Jatropha curcas] |