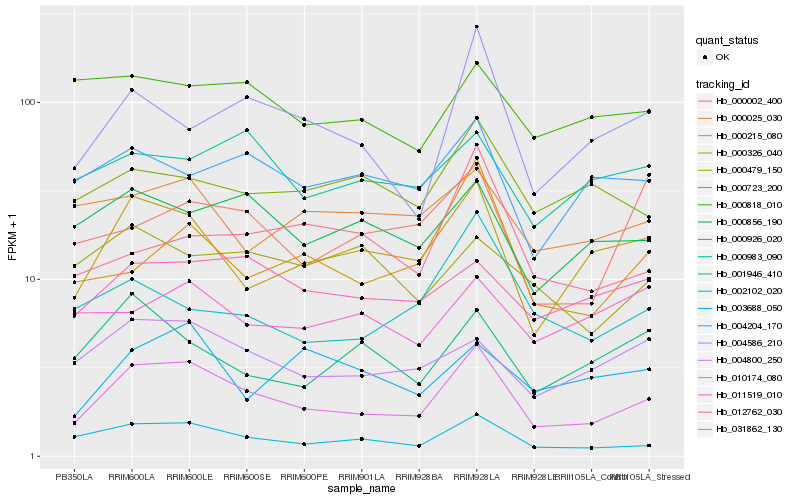
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010174_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_002102_020 |
0.1126389532 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 3 |
Hb_000326_040 |
0.1377255481 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 4 |
Hb_004800_250 |
0.1618447121 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 5 |
Hb_004586_210 |
0.1644539128 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000856_190 |
0.1702464013 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 7 |
Hb_000215_080 |
0.1710389715 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 8 |
Hb_012762_030 |
0.1751031019 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 9 |
Hb_004204_170 |
0.1791721713 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 10 |
Hb_001946_410 |
0.1858635429 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000926_020 |
0.1862024586 |
- |
- |
- |
| 12 |
Hb_000723_200 |
0.1866092372 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000983_090 |
0.1880851397 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 14 |
Hb_000025_030 |
0.1885011407 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000002_400 |
0.1889179197 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor 2 [Jatropha curcas] |
| 16 |
Hb_003688_050 |
0.1897077264 |
- |
- |
PREDICTED: uncharacterized protein LOC105640616 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000818_010 |
0.1912503686 |
- |
- |
Calmodulin-domain protein kinase 5 isoform 1 [Theobroma cacao] |
| 18 |
Hb_031862_130 |
0.1963836118 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_011519_010 |
0.1991498401 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 20 |
Hb_000479_150 |
0.1996684406 |
- |
- |
glycosyl transferase family 8 family protein [Populus trichocarpa] |