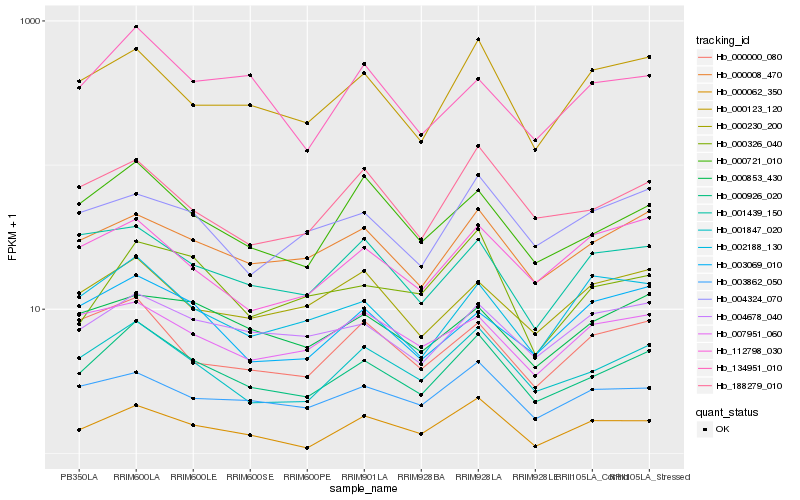
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_001847_020 |
0.0711259481 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Eucalyptus grandis] |
| 3 |
Hb_000721_010 |
0.1017253617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000008_470 |
0.1026257101 |
- |
- |
calcium/calmodulin-dependent protein kinase kinase, putative [Ricinus communis] |
| 5 |
Hb_000123_120 |
0.1075674418 |
- |
- |
actin depolymerizing factor 4 [Hevea brasiliensis] |
| 6 |
Hb_188279_010 |
0.1110532541 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC9-like [Tarenaya hassleriana] |
| 7 |
Hb_003862_050 |
0.1118894522 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 8 |
Hb_004678_040 |
0.1120745984 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000853_430 |
0.1189874307 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 10 |
Hb_000000_080 |
0.1229363932 |
- |
- |
lipoate-protein ligase B, putative [Ricinus communis] |
| 11 |
Hb_134951_010 |
0.1241908173 |
- |
- |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 12 |
Hb_000326_040 |
0.1247550207 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 13 |
Hb_002188_130 |
0.1247655782 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X2 [Jatropha curcas] |
| 14 |
Hb_004324_070 |
0.1255845785 |
- |
- |
protein phosphatase 2C [Hevea brasiliensis] |
| 15 |
Hb_001439_150 |
0.1266262285 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000230_200 |
0.1273906 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 17 |
Hb_000062_350 |
0.1273986784 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_003069_010 |
0.1280110788 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2-like [Jatropha curcas] |
| 19 |
Hb_112798_030 |
0.1286808434 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007951_060 |
0.1289669946 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |