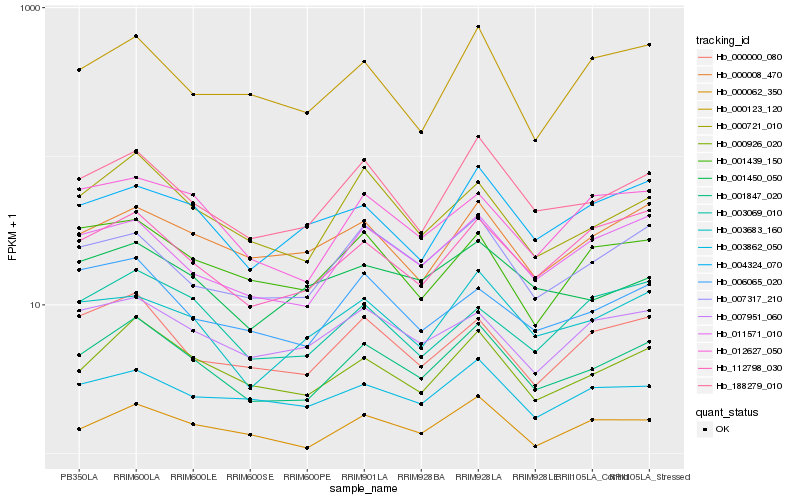
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001847_020 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Eucalyptus grandis] |
| 2 |
Hb_000926_020 |
0.0711259481 |
- |
- |
- |
| 3 |
Hb_188279_010 |
0.077670653 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC9-like [Tarenaya hassleriana] |
| 4 |
Hb_000721_010 |
0.0847585735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011571_010 |
0.0968602735 |
- |
- |
Metal tolerance protein C4, putative [Ricinus communis] |
| 6 |
Hb_000000_080 |
0.1039879021 |
- |
- |
lipoate-protein ligase B, putative [Ricinus communis] |
| 7 |
Hb_003683_160 |
0.1048424316 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 8 |
Hb_007317_210 |
0.105590672 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 9 |
Hb_112798_030 |
0.1056478335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012627_050 |
0.1094575645 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000008_470 |
0.1132864857 |
- |
- |
calcium/calmodulin-dependent protein kinase kinase, putative [Ricinus communis] |
| 12 |
Hb_007951_060 |
0.113845332 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003862_050 |
0.1141006004 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 14 |
Hb_006065_020 |
0.1142539436 |
- |
- |
- |
| 15 |
Hb_004324_070 |
0.1155755139 |
- |
- |
protein phosphatase 2C [Hevea brasiliensis] |
| 16 |
Hb_000123_120 |
0.1163656543 |
- |
- |
actin depolymerizing factor 4 [Hevea brasiliensis] |
| 17 |
Hb_001439_150 |
0.1198579116 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001450_050 |
0.1207489763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000062_350 |
0.1233061838 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_003069_010 |
0.1245797024 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2-like [Jatropha curcas] |