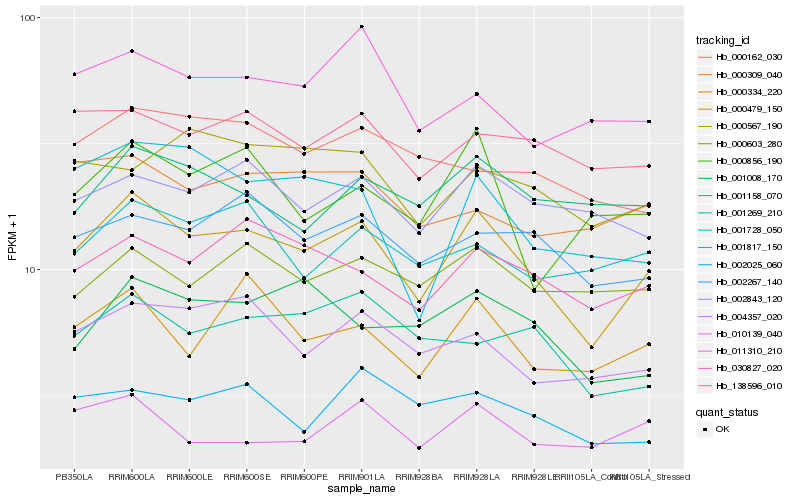
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_150 |
0.0 |
- |
- |
glycosyl transferase family 8 family protein [Populus trichocarpa] |
| 2 |
Hb_030827_020 |
0.102213752 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 3 |
Hb_004357_020 |
0.103305392 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 4 |
Hb_000334_220 |
0.1037046044 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 5 |
Hb_002267_140 |
0.1079946237 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 6 |
Hb_138596_010 |
0.1093509653 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 7 |
Hb_002843_120 |
0.1111002908 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001728_050 |
0.1132023485 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 9 |
Hb_011310_210 |
0.1136559841 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 10 |
Hb_000603_280 |
0.1150024873 |
- |
- |
PREDICTED: uncharacterized protein LOC105638082 [Jatropha curcas] |
| 11 |
Hb_010139_040 |
0.1162270686 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_001817_150 |
0.1163178202 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_002025_060 |
0.1174271871 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g73400, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001269_210 |
0.117475179 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g76280 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000309_040 |
0.1180652644 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 16 |
Hb_000567_190 |
0.1183635266 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001008_170 |
0.1184260103 |
transcription factor |
TF Family: mTERF |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000856_190 |
0.1187778377 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 19 |
Hb_001158_070 |
0.118906643 |
- |
- |
Protein DEK, putative [Ricinus communis] |
| 20 |
Hb_000162_030 |
0.119551412 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |