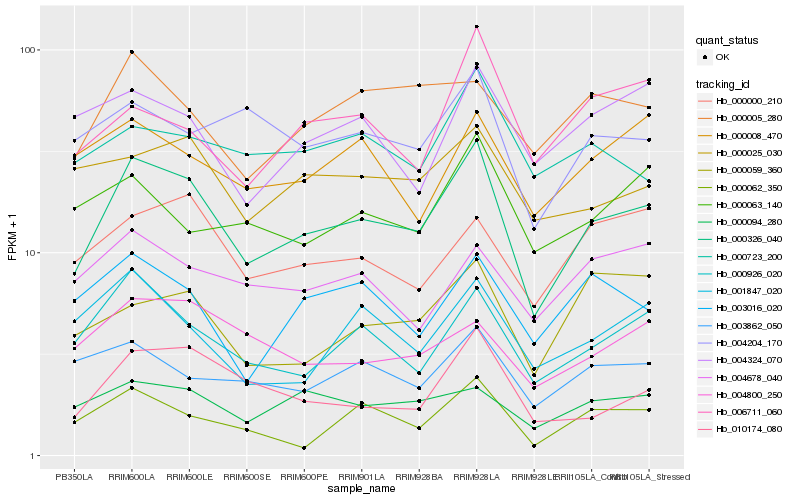
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000326_040 |
0.0 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 2 |
Hb_000926_020 |
0.1247550207 |
- |
- |
- |
| 3 |
Hb_010174_080 |
0.1377255481 |
- |
- |
- |
| 4 |
Hb_000094_280 |
0.1423205637 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 5 |
Hb_003862_050 |
0.1488074912 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 6 |
Hb_000000_210 |
0.1491154907 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000025_030 |
0.1502559224 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_004204_170 |
0.1507928894 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 9 |
Hb_000005_280 |
0.1524104375 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 10 |
Hb_003016_020 |
0.153153697 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000723_200 |
0.1542499561 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004800_250 |
0.1544689761 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 13 |
Hb_001847_020 |
0.1546711499 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Eucalyptus grandis] |
| 14 |
Hb_006711_060 |
0.1568334247 |
- |
- |
Tubulin beta-1 chain [Morus notabilis] |
| 15 |
Hb_000008_470 |
0.1574351351 |
- |
- |
calcium/calmodulin-dependent protein kinase kinase, putative [Ricinus communis] |
| 16 |
Hb_000059_360 |
0.1575944537 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 17 |
Hb_000062_350 |
0.1588473835 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_004324_070 |
0.1591874916 |
- |
- |
protein phosphatase 2C [Hevea brasiliensis] |
| 19 |
Hb_004678_040 |
0.1592259905 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000063_140 |
0.1595085689 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 25 kDa protein [Jatropha curcas] |