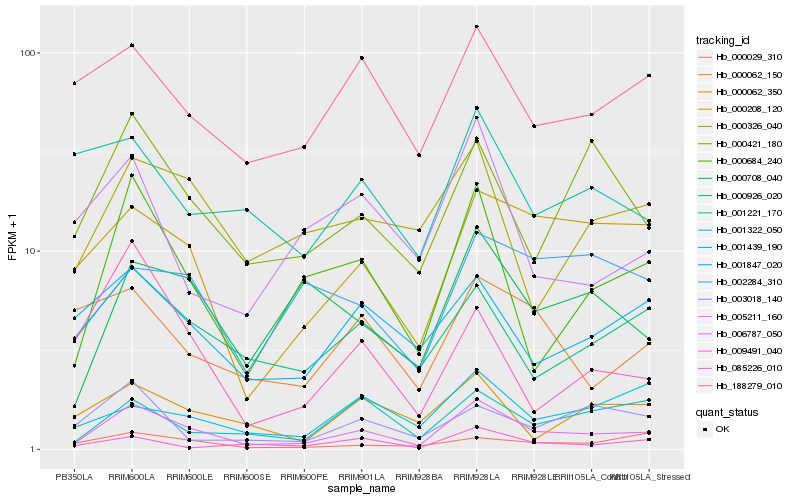
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006787_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_23988 [Jatropha curcas] |
| 2 |
Hb_000684_240 |
0.1713381238 |
- |
- |
hypothetical protein RCOM_0819620 [Ricinus communis] |
| 3 |
Hb_000208_120 |
0.196065319 |
- |
- |
PREDICTED: CAP-Gly domain-containing linker protein 1-like [Jatropha curcas] |
| 4 |
Hb_000421_180 |
0.2030529579 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000926_020 |
0.2063526245 |
- |
- |
- |
| 6 |
Hb_000708_040 |
0.2086804044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001322_050 |
0.2139986519 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001847_020 |
0.2158507024 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Eucalyptus grandis] |
| 9 |
Hb_188279_010 |
0.2168218684 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC9-like [Tarenaya hassleriana] |
| 10 |
Hb_009491_040 |
0.220546759 |
- |
- |
PREDICTED: uncharacterized protein LOC105636967 [Jatropha curcas] |
| 11 |
Hb_000326_040 |
0.2219508305 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 12 |
Hb_000062_350 |
0.2260951817 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000062_150 |
0.2295916273 |
- |
- |
PREDICTED: probable carboxylesterase 16 [Jatropha curcas] |
| 14 |
Hb_001439_190 |
0.2311091791 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 15 |
Hb_000029_310 |
0.2326063167 |
- |
- |
PREDICTED: uncharacterized protein LOC105631832 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005211_160 |
0.2330620364 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 17 |
Hb_003018_140 |
0.2336945312 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 18 |
Hb_001221_170 |
0.234749825 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002284_310 |
0.2352853994 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 20 |
Hb_085226_010 |
0.2356578941 |
- |
- |
hypothetical protein POPTR_0002s03930g [Populus trichocarpa] |