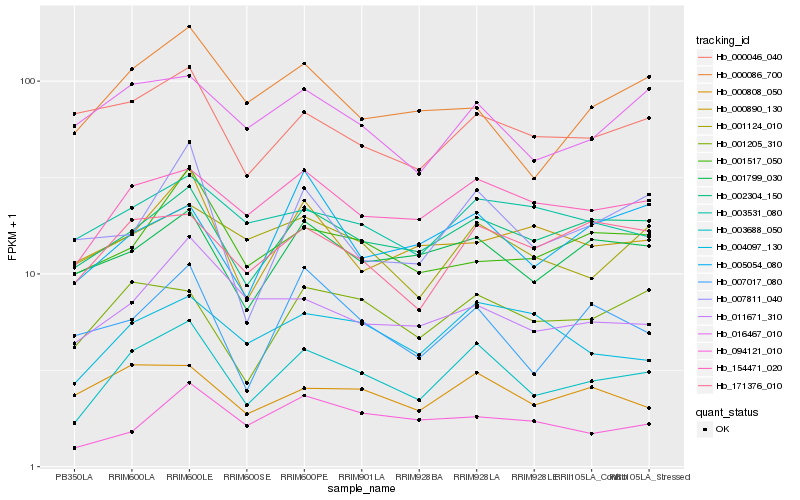
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640616 isoform X2 [Jatropha curcas] |
| 2 |
Hb_154471_020 |
0.1130808039 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |
| 3 |
Hb_002304_150 |
0.1149287458 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001205_310 |
0.1281004152 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL4-like [Jatropha curcas] |
| 5 |
Hb_001799_030 |
0.1290700888 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 6 |
Hb_171376_010 |
0.1319478687 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004097_130 |
0.1327451989 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 8 |
Hb_000808_050 |
0.1365288341 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 9 |
Hb_016467_010 |
0.1374313983 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 10 |
Hb_000086_700 |
0.1385380022 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 11 |
Hb_007017_080 |
0.1393259712 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_011671_310 |
0.1393610125 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 13 |
Hb_001124_010 |
0.1395187649 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007811_040 |
0.1405028688 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 isoform X1 [Gossypium raimondii] |
| 15 |
Hb_001517_050 |
0.1434159601 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000046_040 |
0.1449690399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003531_080 |
0.1453325012 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_094121_010 |
0.146924568 |
- |
- |
- |
| 19 |
Hb_000890_130 |
0.1473456723 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 20 |
Hb_005054_080 |
0.1478166214 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |