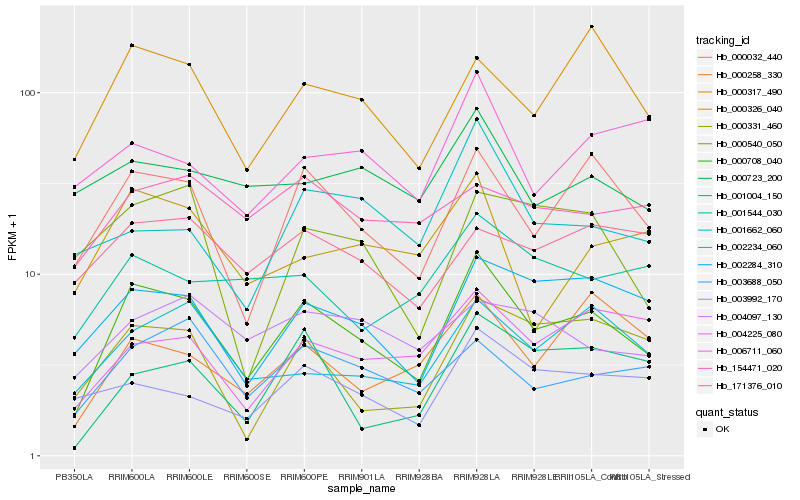
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000708_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000032_440 |
0.1344628669 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_002284_310 |
0.1585759381 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000317_490 |
0.1695810422 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 5 |
Hb_003688_050 |
0.1701103513 |
- |
- |
PREDICTED: uncharacterized protein LOC105640616 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002234_060 |
0.1771221191 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 7 |
Hb_000331_460 |
0.1815683694 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 8 |
Hb_004225_080 |
0.1824744269 |
- |
- |
- |
| 9 |
Hb_003992_170 |
0.1831399879 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 10 |
Hb_001004_150 |
0.1895577369 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 11 |
Hb_001544_030 |
0.1904241238 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C [Jatropha curcas] |
| 12 |
Hb_000326_040 |
0.1915803688 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 13 |
Hb_000258_330 |
0.1916774665 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 14 |
Hb_001662_060 |
0.1918894754 |
- |
- |
SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000540_050 |
0.1947591051 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 16 |
Hb_000723_200 |
0.1959172364 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 17 |
Hb_171376_010 |
0.1966521148 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004097_130 |
0.196962774 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 19 |
Hb_006711_060 |
0.2002789576 |
- |
- |
Tubulin beta-1 chain [Morus notabilis] |
| 20 |
Hb_154471_020 |
0.2018879001 |
- |
- |
squalene synthetase [Euphorbia tirucalli] |