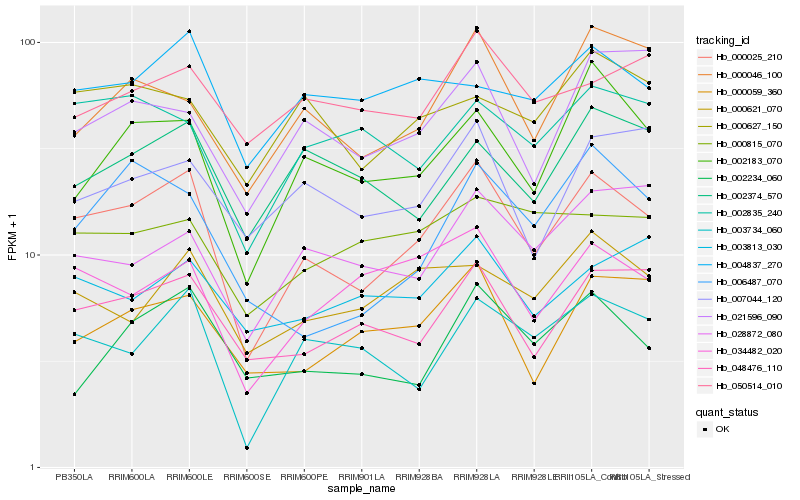
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_210 |
0.0 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_048476_110 |
0.1262699933 |
- |
- |
PREDICTED: adagio protein 3 [Jatropha curcas] |
| 3 |
Hb_003734_060 |
0.1312884653 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002183_070 |
0.1322302841 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_034482_020 |
0.137183949 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000059_360 |
0.1423622507 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 7 |
Hb_000046_100 |
0.1432945929 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000621_070 |
0.1465149008 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 9 |
Hb_002234_060 |
0.1466314081 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 10 |
Hb_002835_240 |
0.1482760203 |
- |
- |
hypothetical protein PRUPE_ppa013861mg [Prunus persica] |
| 11 |
Hb_007044_120 |
0.1501792671 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 12 |
Hb_004837_270 |
0.151459562 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1 [Jatropha curcas] |
| 13 |
Hb_006487_070 |
0.1518296924 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 14 [Jatropha curcas] |
| 14 |
Hb_002374_570 |
0.1519709222 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 15 |
Hb_028872_080 |
0.1528697359 |
- |
- |
PREDICTED: adenylate kinase 4-like [Jatropha curcas] |
| 16 |
Hb_021596_090 |
0.1528768168 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8-B [Jatropha curcas] |
| 17 |
Hb_000627_150 |
0.1529225577 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, cytoplasmic isoform-like [Jatropha curcas] |
| 18 |
Hb_003813_030 |
0.156011365 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 19 |
Hb_000815_070 |
0.1567226346 |
- |
- |
PREDICTED: methyltransferase-like protein 7A [Jatropha curcas] |
| 20 |
Hb_050514_010 |
0.1578887919 |
- |
- |
hypothetical protein 29 [Hevea brasiliensis] |