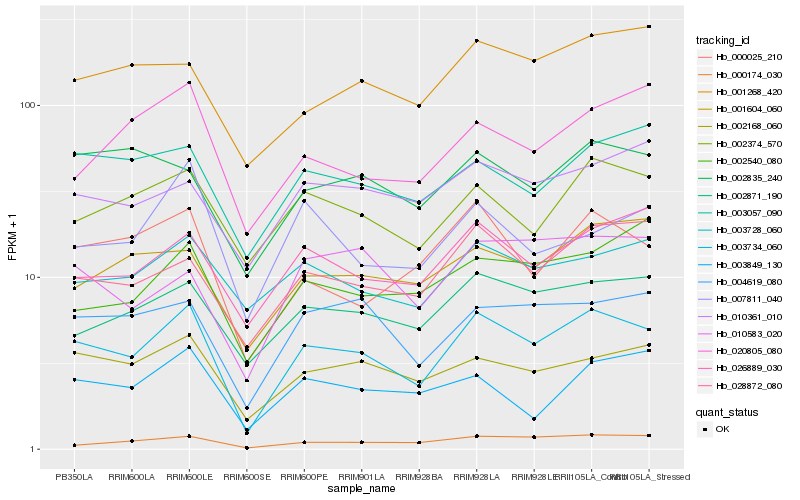
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003734_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002168_060 |
0.1249934208 |
- |
- |
PREDICTED: inositol-pentakisphosphate 2-kinase-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004619_080 |
0.1270095785 |
- |
- |
hypothetical protein PRUPE_ppa011369mg [Prunus persica] |
| 4 |
Hb_028872_080 |
0.1287341513 |
- |
- |
PREDICTED: adenylate kinase 4-like [Jatropha curcas] |
| 5 |
Hb_002871_190 |
0.1290075334 |
- |
- |
PREDICTED: uncharacterized protein LOC105628867 [Jatropha curcas] |
| 6 |
Hb_002374_570 |
0.1310146034 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 7 |
Hb_000025_210 |
0.1312884653 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000174_030 |
0.1325504606 |
- |
- |
hypothetical protein RCOM_1077370 [Ricinus communis] |
| 9 |
Hb_026889_030 |
0.1344531039 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_003728_060 |
0.1400777364 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4-like [Jatropha curcas] |
| 11 |
Hb_003057_090 |
0.142488809 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 12 |
Hb_001604_060 |
0.144751246 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 13 |
Hb_001268_420 |
0.1449886963 |
- |
- |
PREDICTED: uncharacterized protein LOC105630158 [Jatropha curcas] |
| 14 |
Hb_010583_020 |
0.1453855729 |
- |
- |
PREDICTED: uncharacterized protein LOC105646948 [Jatropha curcas] |
| 15 |
Hb_002835_240 |
0.1467963578 |
- |
- |
hypothetical protein PRUPE_ppa013861mg [Prunus persica] |
| 16 |
Hb_010361_010 |
0.1501074086 |
- |
- |
PREDICTED: ras-related protein RABH1b [Jatropha curcas] |
| 17 |
Hb_003849_130 |
0.1503883072 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 18 |
Hb_007811_040 |
0.1527275913 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 isoform X1 [Gossypium raimondii] |
| 19 |
Hb_020805_080 |
0.1534459645 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 20 |
Hb_002540_080 |
0.1535657037 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |