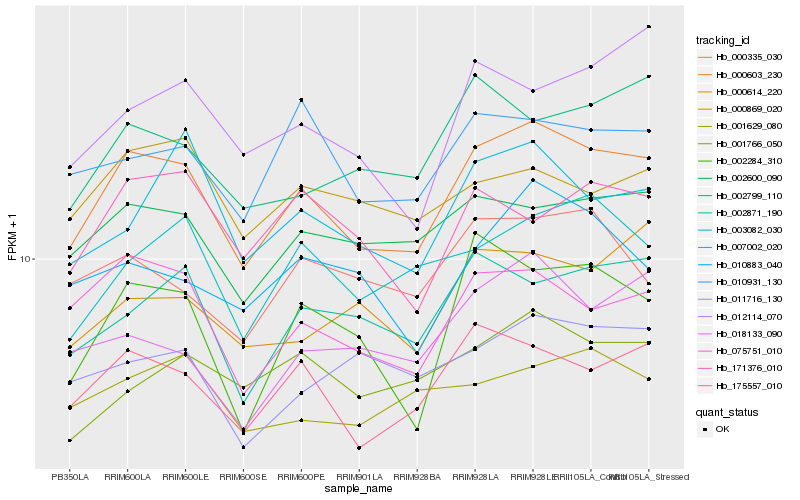
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000603_230 |
0.0 |
- |
- |
copper-transporting atpase paa1, putative [Ricinus communis] |
| 2 |
Hb_175557_010 |
0.0814442089 |
- |
- |
glutamate-1-semialdehyde 2,1-aminomutase, putative [Ricinus communis] |
| 3 |
Hb_075751_010 |
0.0924403493 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002600_090 |
0.0940221439 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_002871_190 |
0.1031731714 |
- |
- |
PREDICTED: uncharacterized protein LOC105628867 [Jatropha curcas] |
| 6 |
Hb_003082_030 |
0.1032036368 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_010931_130 |
0.1044849168 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 8 |
Hb_007002_020 |
0.1090987292 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 9 |
Hb_001766_050 |
0.1101259065 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 10 |
Hb_010883_040 |
0.1108989986 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 11 |
Hb_002284_310 |
0.1109205603 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000335_030 |
0.1115763576 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |
| 13 |
Hb_002799_110 |
0.1116238906 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 14 |
Hb_001629_080 |
0.1131544576 |
- |
- |
hypothetical protein POPTR_0011s12610g [Populus trichocarpa] |
| 15 |
Hb_171376_010 |
0.1131595077 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 16 |
Hb_018133_090 |
0.1149419329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011716_130 |
0.1150001203 |
- |
- |
PREDICTED: probable acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, mitochondrial isoform X5 [Populus euphratica] |
| 18 |
Hb_000869_020 |
0.1154548084 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 19 |
Hb_012114_070 |
0.1156655881 |
- |
- |
PREDICTED: uncharacterized protein LOC105645812 [Jatropha curcas] |
| 20 |
Hb_000614_220 |
0.1164774114 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |