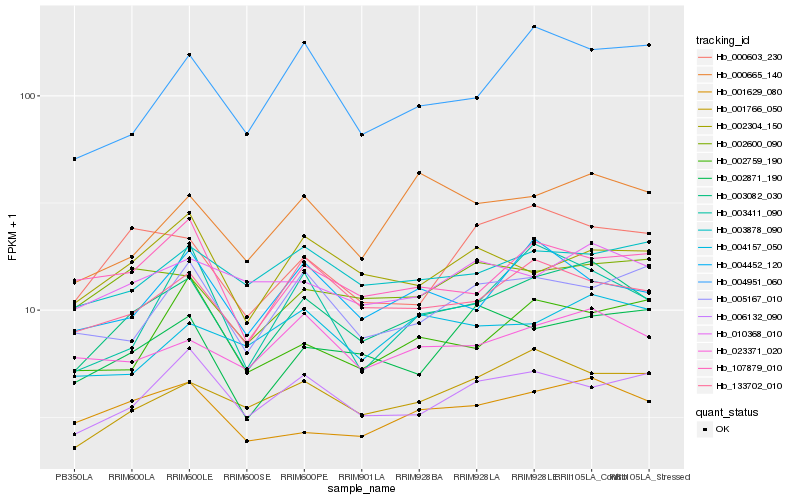
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003082_030 |
0.0 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001629_080 |
0.091631411 |
- |
- |
hypothetical protein POPTR_0011s12610g [Populus trichocarpa] |
| 3 |
Hb_133702_010 |
0.0933360037 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_001766_050 |
0.094026451 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 5 |
Hb_023371_020 |
0.1006139653 |
- |
- |
PREDICTED: uncharacterized protein LOC105633512 [Jatropha curcas] |
| 6 |
Hb_006132_090 |
0.1012307911 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000665_140 |
0.1018681229 |
- |
- |
PREDICTED: protein MEMO1 [Jatropha curcas] |
| 8 |
Hb_000603_230 |
0.1032036368 |
- |
- |
copper-transporting atpase paa1, putative [Ricinus communis] |
| 9 |
Hb_004452_120 |
0.1037673226 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 10 |
Hb_002600_090 |
0.1042384152 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_002871_190 |
0.1054322843 |
- |
- |
PREDICTED: uncharacterized protein LOC105628867 [Jatropha curcas] |
| 12 |
Hb_003878_090 |
0.1056765859 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 13 |
Hb_004951_060 |
0.1068180403 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 14 |
Hb_005167_010 |
0.1085563193 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 15 |
Hb_010368_010 |
0.1095862333 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 16 |
Hb_107879_010 |
0.1096232917 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 17 |
Hb_003411_090 |
0.1097033513 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_002304_150 |
0.1104061804 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_004157_050 |
0.1116182642 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002759_190 |
0.1124351974 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |