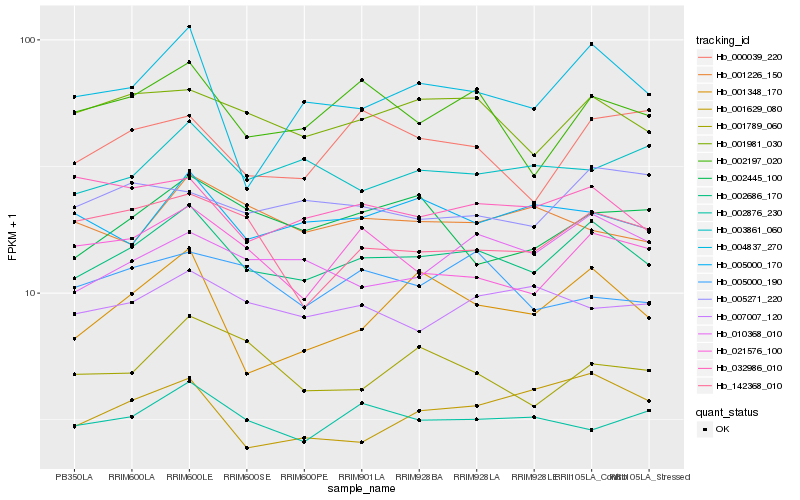
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001981_030 |
0.0686507606 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_002197_020 |
0.0779935546 |
- |
- |
DNA-directed RNA polymerase family protein [Theobroma cacao] |
| 4 |
Hb_010368_010 |
0.0801597589 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 5 |
Hb_002876_230 |
0.082628339 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 6 |
Hb_021576_100 |
0.0826731689 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 7 |
Hb_005000_170 |
0.0850690362 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 8 |
Hb_007007_120 |
0.0851187194 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004837_270 |
0.0853391003 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1 [Jatropha curcas] |
| 10 |
Hb_003861_060 |
0.0857047619 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 11 |
Hb_032986_010 |
0.0863089916 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 12 |
Hb_001348_170 |
0.0885953592 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 13 |
Hb_005271_220 |
0.0892484325 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |
| 14 |
Hb_142368_010 |
0.0901939498 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002445_100 |
0.0902969988 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001629_080 |
0.0912648294 |
- |
- |
hypothetical protein POPTR_0011s12610g [Populus trichocarpa] |
| 17 |
Hb_000039_220 |
0.0929781467 |
- |
- |
PREDICTED: vesicle-associated protein 4-2-like [Gossypium raimondii] |
| 18 |
Hb_001789_060 |
0.0935316485 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 19 |
Hb_001226_150 |
0.0938741907 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_005000_190 |
0.0941047729 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |