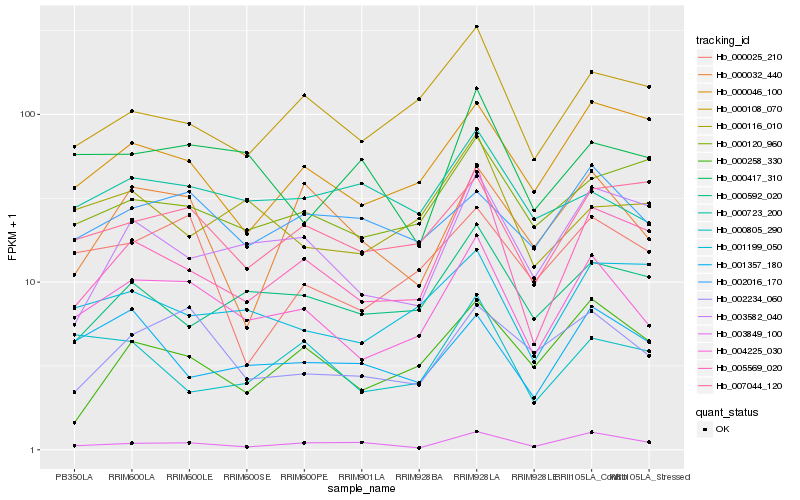
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004225_030 |
0.0 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 2 |
Hb_002234_060 |
0.1486452876 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 3 |
Hb_005569_020 |
0.1503797836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002016_170 |
0.1592312823 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000046_100 |
0.162032174 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000108_070 |
0.1625227791 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 7 |
Hb_000025_210 |
0.1637829352 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001199_050 |
0.1669561719 |
- |
- |
PREDICTED: DNA-3-methyladenine glycosylase-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_007044_120 |
0.1671071961 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 10 |
Hb_000592_020 |
0.1675692892 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_000723_200 |
0.1684231061 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003582_040 |
0.1685812637 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 13 |
Hb_003849_100 |
0.1693003051 |
- |
- |
PREDICTED: putative transcription elongation factor SPT5 homolog 1 [Populus euphratica] |
| 14 |
Hb_001357_180 |
0.1707516693 |
- |
- |
hypothetical protein CISIN_1g036415mg [Citrus sinensis] |
| 15 |
Hb_000116_010 |
0.1708943957 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000120_960 |
0.17150528 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000032_440 |
0.1735703571 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000258_330 |
0.1754201786 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 19 |
Hb_000417_310 |
0.1757088375 |
- |
- |
uncharacterized protein LOC100272531 [Zea mays] |
| 20 |
Hb_000805_290 |
0.177004054 |
- |
- |
PREDICTED: cytochrome P450 710A1-like [Jatropha curcas] |