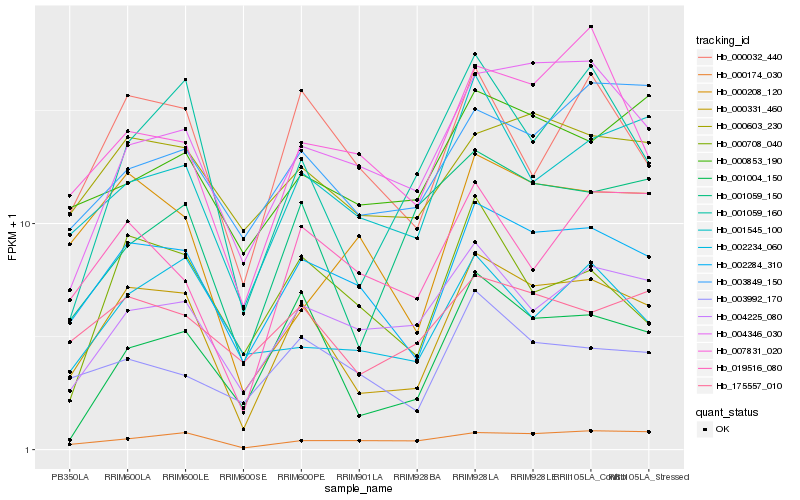
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_460 |
0.0 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 2 |
Hb_002284_310 |
0.1126821909 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001004_150 |
0.1542149078 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 4 |
Hb_000032_440 |
0.1573239476 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_001545_100 |
0.1587359599 |
- |
- |
PREDICTED: probable F-actin-capping protein subunit beta [Vitis vinifera] |
| 6 |
Hb_000603_230 |
0.1616125665 |
- |
- |
copper-transporting atpase paa1, putative [Ricinus communis] |
| 7 |
Hb_004225_080 |
0.1640964095 |
- |
- |
- |
| 8 |
Hb_001059_160 |
0.1654566747 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 9 |
Hb_004346_030 |
0.1659873248 |
- |
- |
sucrose phosphate phosphatase, putative [Ricinus communis] |
| 10 |
Hb_175557_010 |
0.1662710539 |
- |
- |
glutamate-1-semialdehyde 2,1-aminomutase, putative [Ricinus communis] |
| 11 |
Hb_000174_030 |
0.1680327245 |
- |
- |
hypothetical protein RCOM_1077370 [Ricinus communis] |
| 12 |
Hb_001059_150 |
0.1702410827 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 13 |
Hb_019516_080 |
0.1747597546 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003992_170 |
0.1752282211 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 15 |
Hb_003849_150 |
0.1801480997 |
- |
- |
PREDICTED: uncharacterized protein LOC105644698 [Jatropha curcas] |
| 16 |
Hb_007831_020 |
0.1809313681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000708_040 |
0.1815683694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002234_060 |
0.1835488588 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 19 |
Hb_000853_190 |
0.183651689 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 20 |
Hb_000208_120 |
0.1838509261 |
- |
- |
PREDICTED: CAP-Gly domain-containing linker protein 1-like [Jatropha curcas] |