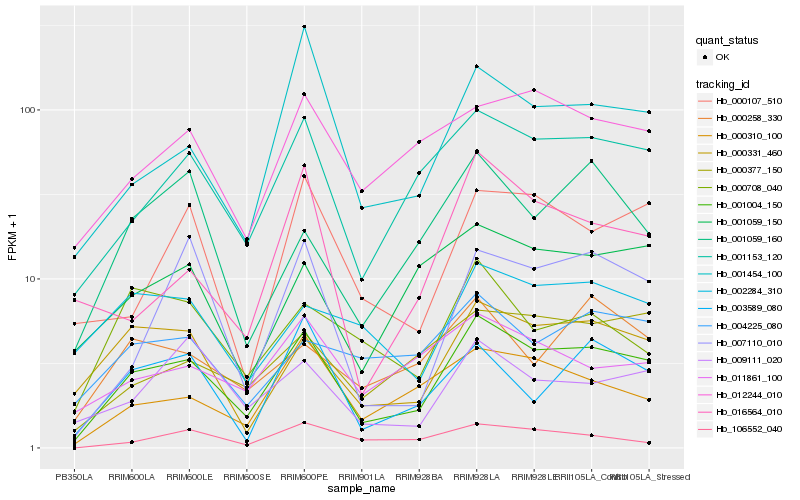
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001004_150 |
0.0 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 2 |
Hb_001153_120 |
0.1272001209 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 3 |
Hb_000331_460 |
0.1542149078 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 4 |
Hb_000310_100 |
0.1674090274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001454_100 |
0.1730800898 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 6 |
Hb_012244_010 |
0.17357924 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 7 |
Hb_000377_150 |
0.1770964429 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 8 |
Hb_001059_150 |
0.1778373314 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 9 |
Hb_000107_510 |
0.1784777678 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 10 |
Hb_009111_020 |
0.1792534565 |
- |
- |
PREDICTED: calcium-dependent protein kinase 20-like [Jatropha curcas] |
| 11 |
Hb_011861_100 |
0.1797642522 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_003589_080 |
0.1824968879 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 13 |
Hb_007110_010 |
0.1834934359 |
- |
- |
PREDICTED: accessory gland protein Acp36DE [Jatropha curcas] |
| 14 |
Hb_004225_080 |
0.1867523885 |
- |
- |
- |
| 15 |
Hb_000258_330 |
0.1880137443 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_000708_040 |
0.1895577369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002284_310 |
0.1905339782 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 18 |
Hb_106552_040 |
0.1906825173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_016564_010 |
0.1908403971 |
- |
- |
PREDICTED: uncharacterized protein LOC101764501 [Setaria italica] |
| 20 |
Hb_001059_160 |
0.1919682983 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |