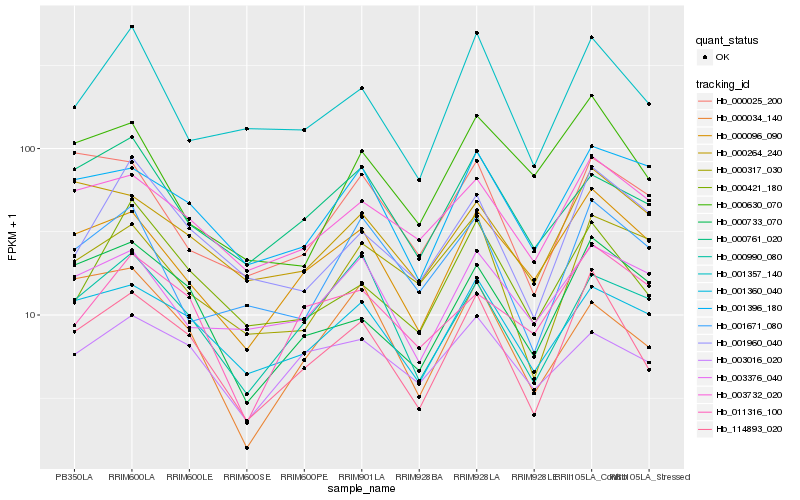
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114893_020 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000096_090 |
0.130567032 |
- |
- |
hypothetical protein JCGZ_12551 [Jatropha curcas] |
| 3 |
Hb_000733_070 |
0.1367426125 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 4 |
Hb_001360_040 |
0.1380417868 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001357_140 |
0.1432816871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000421_180 |
0.1435550404 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_003732_020 |
0.1458827668 |
- |
- |
PREDICTED: phosphoglucomutase, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_001960_040 |
0.1525245497 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 9 |
Hb_001396_180 |
0.1544642594 |
- |
- |
PREDICTED: uncharacterized protein LOC105644834 [Jatropha curcas] |
| 10 |
Hb_000264_240 |
0.1567593957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000990_080 |
0.1607841516 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 12 |
Hb_003016_020 |
0.1636776458 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000630_070 |
0.1646771342 |
- |
- |
PREDICTED: cell division protein FtsZ homolog 2-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000761_020 |
0.1651904064 |
- |
- |
PREDICTED: uncharacterized protein LOC105638198 [Jatropha curcas] |
| 15 |
Hb_001671_080 |
0.1653510843 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000025_200 |
0.1671695032 |
- |
- |
PREDICTED: cell division control protein 2 homolog A isoform X1 [Jatropha curcas] |
| 17 |
Hb_000317_030 |
0.1675952534 |
transcription factor |
TF Family: ULT |
PREDICTED: protein ULTRAPETALA 1-like [Jatropha curcas] |
| 18 |
Hb_003376_040 |
0.1679552772 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 19 |
Hb_000034_140 |
0.1681634909 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_011316_100 |
0.1683117635 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4-like [Jatropha curcas] |