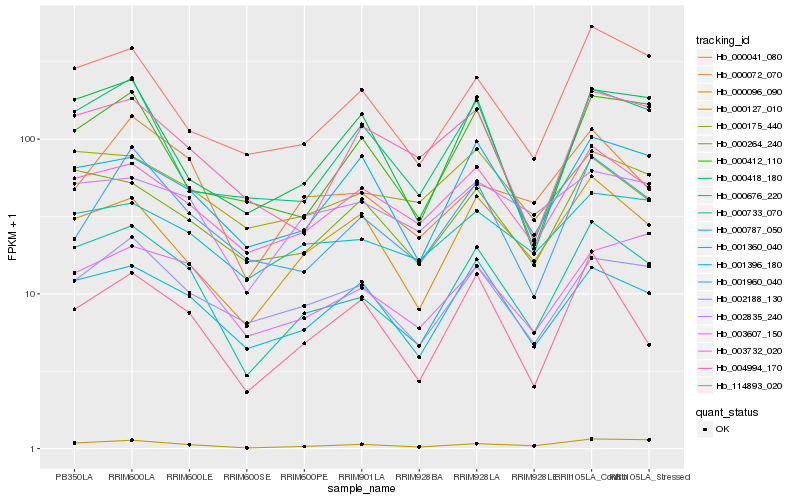
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000733_070 |
0.0 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 2 |
Hb_000127_010 |
0.1202581917 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Populus euphratica] |
| 3 |
Hb_000264_240 |
0.1204324664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003732_020 |
0.1225902797 |
- |
- |
PREDICTED: phosphoglucomutase, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_000041_080 |
0.1258707863 |
- |
- |
PREDICTED: universal stress protein A-like protein [Jatropha curcas] |
| 6 |
Hb_001360_040 |
0.1272580916 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000096_090 |
0.1304800201 |
- |
- |
hypothetical protein JCGZ_12551 [Jatropha curcas] |
| 8 |
Hb_002188_130 |
0.1317978827 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X2 [Jatropha curcas] |
| 9 |
Hb_114893_020 |
0.1367426125 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000072_070 |
0.1414418418 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl diphosphate synthase [Hevea brasiliensis] |
| 11 |
Hb_001396_180 |
0.141522153 |
- |
- |
PREDICTED: uncharacterized protein LOC105644834 [Jatropha curcas] |
| 12 |
Hb_003607_150 |
0.1455713263 |
- |
- |
PREDICTED: thioredoxin-like protein 4B [Jatropha curcas] |
| 13 |
Hb_001960_040 |
0.145624695 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 14 |
Hb_000418_180 |
0.1459210391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000175_440 |
0.1470520623 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 16 |
Hb_000676_220 |
0.147490937 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 17 |
Hb_000412_110 |
0.147859211 |
- |
- |
Rop4 [Hevea brasiliensis] |
| 18 |
Hb_000787_050 |
0.1488130135 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_002835_240 |
0.1495597791 |
- |
- |
hypothetical protein PRUPE_ppa013861mg [Prunus persica] |
| 20 |
Hb_004994_170 |
0.1499064821 |
- |
- |
PREDICTED: uncharacterized protein LOC105643024 [Jatropha curcas] |