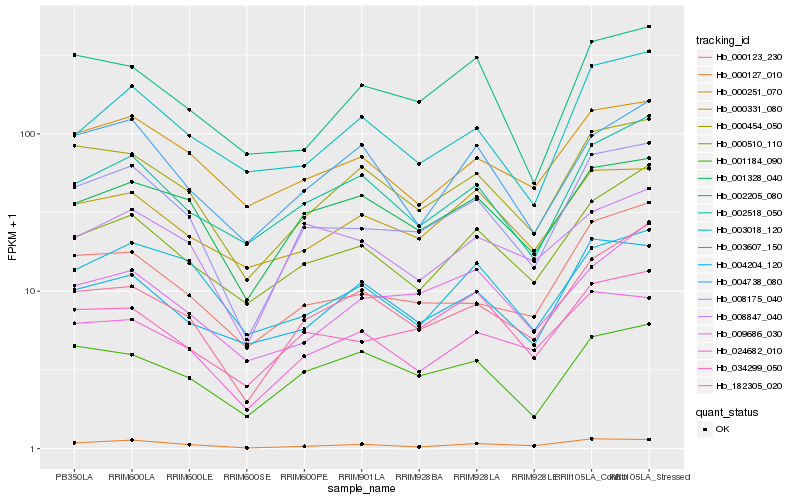
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008175_040 |
0.0 |
- |
- |
PREDICTED: sm-like protein LSM36B [Jatropha curcas] |
| 2 |
Hb_000454_050 |
0.0877225906 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 [Vitis vinifera] |
| 3 |
Hb_000127_010 |
0.0939667531 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Populus euphratica] |
| 4 |
Hb_000123_230 |
0.0996108794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001328_040 |
0.1064099052 |
- |
- |
PREDICTED: uncharacterized protein LOC105636146 [Jatropha curcas] |
| 6 |
Hb_182305_020 |
0.1064448091 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_000510_110 |
0.1073558925 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |
| 8 |
Hb_034299_050 |
0.1084977129 |
- |
- |
hypothetical protein CICLE_v10031248mg [Citrus clementina] |
| 9 |
Hb_024682_010 |
0.1093446424 |
- |
- |
sterol isomerase, putative [Ricinus communis] |
| 10 |
Hb_000251_070 |
0.1100199328 |
- |
- |
hypothetical protein L484_010641 [Morus notabilis] |
| 11 |
Hb_002518_050 |
0.1122162763 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM10 [Jatropha curcas] |
| 12 |
Hb_001184_090 |
0.1195683392 |
- |
- |
PREDICTED: TIP41-like protein [Jatropha curcas] |
| 13 |
Hb_003607_150 |
0.1201642478 |
- |
- |
PREDICTED: thioredoxin-like protein 4B [Jatropha curcas] |
| 14 |
Hb_000331_080 |
0.1221165843 |
- |
- |
PREDICTED: UPF0235 protein At5g63440 isoform X1 [Gossypium raimondii] |
| 15 |
Hb_004738_080 |
0.1223442159 |
- |
- |
transcription initiation factor TFIID subunit 13-like [Jatropha curcas] |
| 16 |
Hb_004204_120 |
0.1231123901 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 17 |
Hb_008847_040 |
0.1242710341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002205_080 |
0.1246180731 |
- |
- |
40S ribosomal protein S26, putative [Ricinus communis] |
| 19 |
Hb_009686_030 |
0.1250467416 |
- |
- |
- |
| 20 |
Hb_003018_120 |
0.1250633124 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein F [Jatropha curcas] |