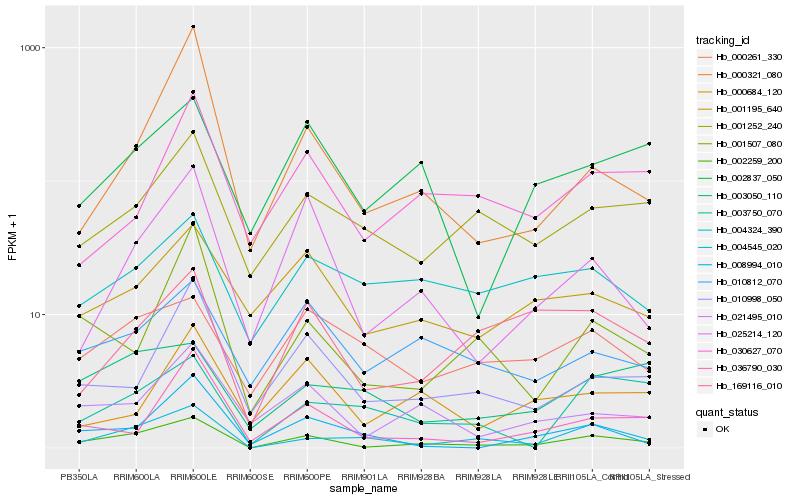
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004545_020 |
0.2079185459 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 3 |
Hb_021495_010 |
0.2129614361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_025214_120 |
0.2257654122 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 5 |
Hb_010998_050 |
0.2299424211 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 6 |
Hb_010812_070 |
0.2373975731 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: protein NETWORKED 4A [Jatropha curcas] |
| 7 |
Hb_001252_240 |
0.2394563463 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 8 |
Hb_001507_080 |
0.2412102177 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 9 |
Hb_001195_640 |
0.2430827724 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 10 |
Hb_036790_030 |
0.2457149467 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 11 |
Hb_002837_050 |
0.2463762639 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 12 |
Hb_169116_010 |
0.2499328686 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_030627_070 |
0.2548700901 |
- |
- |
histone H4 [Zea mays] |
| 14 |
Hb_000321_080 |
0.257016167 |
- |
- |
histone H4 [Zea mays] |
| 15 |
Hb_000261_330 |
0.2593030558 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Amborella trichopoda] |
| 16 |
Hb_003050_110 |
0.2617297354 |
- |
- |
PREDICTED: prefoldin subunit 6 [Populus euphratica] |
| 17 |
Hb_000684_120 |
0.2635619273 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004324_390 |
0.2641597444 |
- |
- |
PREDICTED: plastidial pyruvate kinase 2 [Jatropha curcas] |
| 19 |
Hb_003750_070 |
0.264944523 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_008994_010 |
0.2651047203 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |