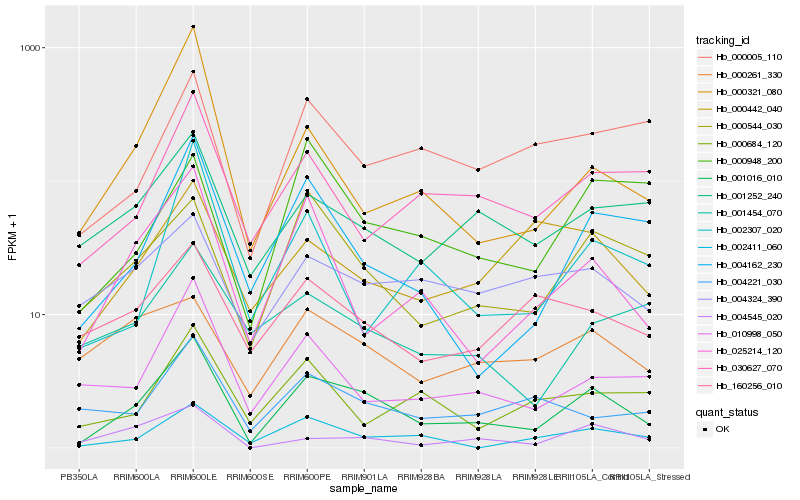
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001016_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004162_230 |
0.2078143983 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 3 |
Hb_000544_030 |
0.2120861359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002411_060 |
0.2132260675 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 5 |
Hb_025214_120 |
0.2162017469 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 6 |
Hb_010998_050 |
0.2236584246 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 7 |
Hb_030627_070 |
0.2253404789 |
- |
- |
histone H4 [Zea mays] |
| 8 |
Hb_001252_240 |
0.2263129473 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 9 |
Hb_004545_020 |
0.2280836642 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 10 |
Hb_004221_030 |
0.2395068526 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000005_110 |
0.2406970381 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 12 |
Hb_004324_390 |
0.2422207526 |
- |
- |
PREDICTED: plastidial pyruvate kinase 2 [Jatropha curcas] |
| 13 |
Hb_002307_020 |
0.2438123781 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 14 |
Hb_160256_010 |
0.2448309717 |
- |
- |
- |
| 15 |
Hb_000321_080 |
0.2450705575 |
- |
- |
histone H4 [Zea mays] |
| 16 |
Hb_000684_120 |
0.248163022 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000442_040 |
0.2486166615 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000261_330 |
0.2487770056 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Amborella trichopoda] |
| 19 |
Hb_001454_070 |
0.2494172213 |
- |
- |
Uveal autoantigen with coiled-coil domains and ankyrin repeats isoform 1 [Theobroma cacao] |
| 20 |
Hb_000948_200 |
0.2518970716 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |