| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
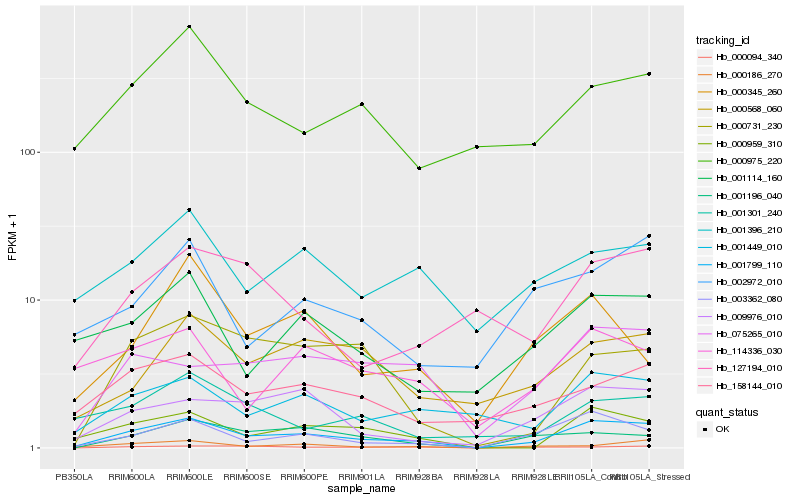
Hb_001799_110 |
0.0 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 2 |
Hb_003362_080 |
0.1720397787 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 3 |
Hb_000959_310 |
0.1871294764 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 4 |
Hb_075265_010 |
0.1938997787 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_009976_010 |
0.1946999972 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 6 |
Hb_001449_010 |
0.1958401213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_158144_010 |
0.2042215604 |
- |
- |
- |
| 8 |
Hb_000186_270 |
0.2090269795 |
- |
- |
hypothetical protein JCGZ_02112 [Jatropha curcas] |
| 9 |
Hb_001114_160 |
0.2111223069 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000568_060 |
0.2132299157 |
- |
- |
PREDICTED: uncharacterized protein LOC105650906 [Jatropha curcas] |
| 11 |
Hb_001196_040 |
0.2141910399 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 12 |
Hb_000094_340 |
0.2175651101 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 13 |
Hb_001396_210 |
0.2206425363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000731_230 |
0.222544443 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 15 |
Hb_000975_220 |
0.2231175277 |
- |
- |
Histone H2B [Medicago truncatula] |
| 16 |
Hb_114336_030 |
0.2240946128 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 17 |
Hb_002972_010 |
0.2251668308 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 18 |
Hb_001301_240 |
0.2290084424 |
- |
- |
- |
| 19 |
Hb_000345_260 |
0.2329788741 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_127194_010 |
0.2340170963 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |