| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
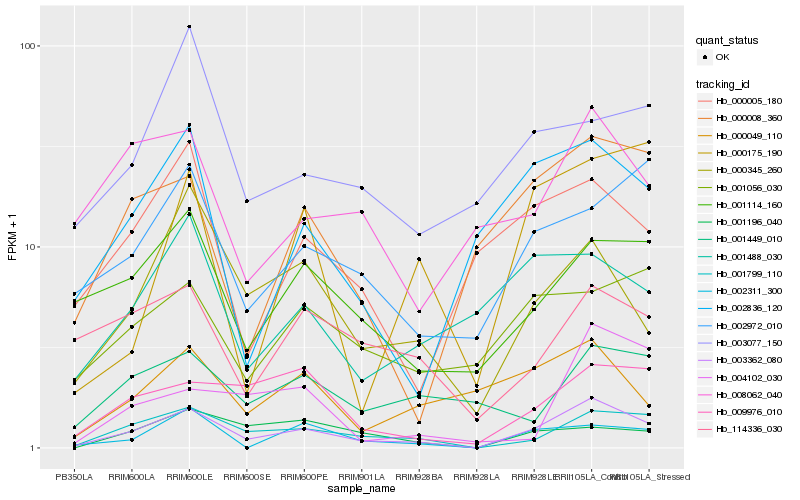
Hb_003362_080 |
0.0 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 2 |
Hb_001799_110 |
0.1720397787 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_000345_260 |
0.2206780094 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_009976_010 |
0.2270248822 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 5 |
Hb_001114_160 |
0.2287784819 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002311_300 |
0.2326322691 |
- |
- |
PREDICTED: J protein JJJ2 [Jatropha curcas] |
| 7 |
Hb_002972_010 |
0.242452822 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 8 |
Hb_002836_120 |
0.2461975595 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor At1g16060 isoform X1 [Populus euphratica] |
| 9 |
Hb_000008_360 |
0.2497562859 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 10 |
Hb_001056_030 |
0.2512048582 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 11 |
Hb_114336_030 |
0.2512817648 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |
| 12 |
Hb_001488_030 |
0.2524891609 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 13 |
Hb_000049_110 |
0.2537358274 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 14 |
Hb_008062_040 |
0.255839413 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001449_010 |
0.2564953935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000005_180 |
0.2566787067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001196_040 |
0.2584187199 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 18 |
Hb_003077_150 |
0.264904304 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 19 |
Hb_004102_030 |
0.268715502 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 20 |
Hb_000175_190 |
0.2690122114 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |