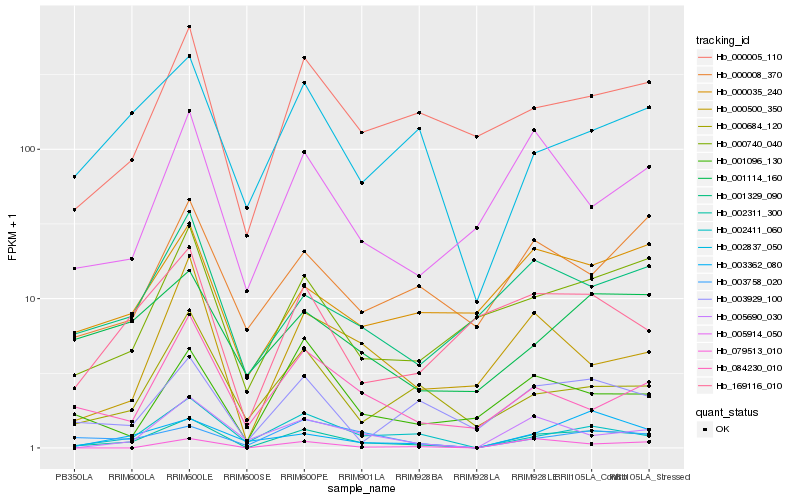
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_300 |
0.0 |
- |
- |
PREDICTED: J protein JJJ2 [Jatropha curcas] |
| 2 |
Hb_002411_060 |
0.2087521981 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 3 |
Hb_003929_100 |
0.2196606458 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Populus euphratica] |
| 4 |
Hb_000740_040 |
0.2210198542 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 5 |
Hb_000005_110 |
0.2238392056 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 6 |
Hb_005690_030 |
0.2304789449 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 7 |
Hb_003362_080 |
0.2326322691 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 8 |
Hb_000500_350 |
0.2358134747 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 9 |
Hb_002837_050 |
0.2374926621 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 10 |
Hb_005914_050 |
0.2401157349 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001329_090 |
0.2408177889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000684_120 |
0.2415249917 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003758_020 |
0.243235693 |
- |
- |
UGTPg34 [Panax ginseng] |
| 14 |
Hb_001096_130 |
0.2440014728 |
- |
- |
PREDICTED: RING-H2 finger protein ATL7-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_169116_010 |
0.2444140829 |
- |
- |
PREDICTED: fatty-acid-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_000035_240 |
0.2454257594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001114_160 |
0.2460674649 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000008_370 |
0.2460733188 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 19 |
Hb_084230_010 |
0.246943372 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 20 |
Hb_079513_010 |
0.2477152919 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |